Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for LEF1
Z-value: 2.41
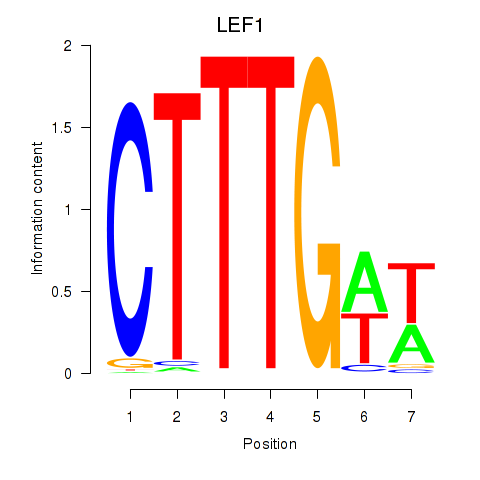
Transcription factors associated with LEF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LEF1
|
ENSG00000138795.5 | LEF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LEF1 | hg19_v2_chr4_-_109087872_109087881, hg19_v2_chr4_-_109088940_109089037 | -0.08 | 8.5e-01 | Click! |
Activity profile of LEF1 motif
Sorted Z-values of LEF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LEF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_29027696 | 3.62 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr16_+_68679193 | 2.83 |
ENST00000581171.1 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr16_+_68678892 | 2.54 |
ENST00000429102.2 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr9_-_23825956 | 2.41 |
ENST00000397312.2 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr16_+_68678739 | 2.34 |
ENST00000264012.4 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr8_-_42234745 | 2.13 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chr15_+_43885252 | 2.11 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr1_+_82266053 | 2.11 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr15_+_43985725 | 2.06 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr5_+_66124590 | 2.05 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr15_+_43985084 | 2.04 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr7_-_41742697 | 1.83 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr2_-_161056802 | 1.75 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr2_-_161056762 | 1.75 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr14_-_23624511 | 1.68 |
ENST00000529705.2 |
SLC7A8 |
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr9_-_23826298 | 1.64 |
ENST00000380117.1 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr3_+_189349162 | 1.63 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chrX_-_32173579 | 1.61 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr2_-_165424973 | 1.51 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr19_-_6690723 | 1.48 |
ENST00000601008.1 |
C3 |
complement component 3 |
| chr7_+_69064300 | 1.45 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr6_-_11807277 | 1.39 |
ENST00000379415.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr15_+_41136216 | 1.29 |
ENST00000562057.1 ENST00000344051.4 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr11_-_87908600 | 1.27 |
ENST00000531138.1 ENST00000526372.1 ENST00000243662.6 |
RAB38 |
RAB38, member RAS oncogene family |
| chr6_+_37137939 | 1.27 |
ENST00000373509.5 |
PIM1 |
pim-1 oncogene |
| chr5_+_145317356 | 1.24 |
ENST00000511217.1 |
SH3RF2 |
SH3 domain containing ring finger 2 |
| chr8_-_15095832 | 1.24 |
ENST00000382080.1 |
SGCZ |
sarcoglycan, zeta |
| chr19_+_751122 | 1.18 |
ENST00000215582.6 |
MISP |
mitotic spindle positioning |
| chr8_-_21988558 | 1.14 |
ENST00000312841.8 |
HR |
hair growth associated |
| chr6_-_11779403 | 1.13 |
ENST00000414691.3 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr5_-_175964366 | 1.13 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr1_-_116383322 | 1.13 |
ENST00000429731.1 |
NHLH2 |
nescient helix loop helix 2 |
| chr12_-_31477072 | 1.12 |
ENST00000454658.2 |
FAM60A |
family with sequence similarity 60, member A |
| chr7_-_22233442 | 1.11 |
ENST00000401957.2 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_+_85360499 | 1.09 |
ENST00000282111.3 |
TCF7L1 |
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr6_-_11779174 | 1.08 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr17_-_39507064 | 1.08 |
ENST00000007735.3 |
KRT33A |
keratin 33A |
| chr12_-_67072714 | 1.05 |
ENST00000545666.1 ENST00000398016.3 ENST00000359742.4 ENST00000286445.7 ENST00000538211.1 |
GRIP1 |
glutamate receptor interacting protein 1 |
| chr16_+_57673430 | 1.04 |
ENST00000540164.2 ENST00000568531.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr7_+_16793160 | 1.03 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr8_-_80993010 | 1.03 |
ENST00000537855.1 ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52 |
tumor protein D52 |
| chr7_+_20370300 | 1.01 |
ENST00000537992.1 |
ITGB8 |
integrin, beta 8 |
| chr3_-_56809685 | 0.99 |
ENST00000413728.2 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr10_-_116164450 | 0.98 |
ENST00000369271.3 |
AFAP1L2 |
actin filament associated protein 1-like 2 |
| chr18_-_53089723 | 0.98 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr1_-_116383738 | 0.97 |
ENST00000320238.3 |
NHLH2 |
nescient helix loop helix 2 |
| chr7_+_20370746 | 0.96 |
ENST00000222573.4 |
ITGB8 |
integrin, beta 8 |
| chr5_+_140248518 | 0.95 |
ENST00000398640.2 |
PCDHA11 |
protocadherin alpha 11 |
| chr10_-_116164239 | 0.95 |
ENST00000419268.1 ENST00000304129.4 ENST00000545353.1 |
AFAP1L2 |
actin filament associated protein 1-like 2 |
| chr9_+_133971909 | 0.94 |
ENST00000247291.3 ENST00000372302.1 ENST00000372300.1 ENST00000372298.1 |
AIF1L |
allograft inflammatory factor 1-like |
| chr9_+_133971863 | 0.91 |
ENST00000372309.3 |
AIF1L |
allograft inflammatory factor 1-like |
| chr9_-_117150243 | 0.90 |
ENST00000374088.3 |
AKNA |
AT-hook transcription factor |
| chr18_-_52989217 | 0.90 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr12_-_95611149 | 0.89 |
ENST00000549499.1 ENST00000343958.4 ENST00000546711.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr2_+_171571827 | 0.88 |
ENST00000375281.3 |
SP5 |
Sp5 transcription factor |
| chr16_+_57673207 | 0.88 |
ENST00000564783.1 ENST00000564729.1 ENST00000565976.1 ENST00000566508.1 ENST00000544297.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr6_-_11779840 | 0.85 |
ENST00000506810.1 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr18_+_55888767 | 0.85 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr10_-_116444371 | 0.84 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr5_+_147258266 | 0.82 |
ENST00000296694.4 |
SCGB3A2 |
secretoglobin, family 3A, member 2 |
| chr18_-_74207146 | 0.79 |
ENST00000443185.2 |
ZNF516 |
zinc finger protein 516 |
| chr14_+_75746781 | 0.79 |
ENST00000555347.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr2_+_18059906 | 0.78 |
ENST00000304101.4 |
KCNS3 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr8_-_17579726 | 0.78 |
ENST00000381861.3 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr1_-_32801825 | 0.78 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr5_+_145316120 | 0.76 |
ENST00000359120.4 |
SH3RF2 |
SH3 domain containing ring finger 2 |
| chr11_-_94965667 | 0.73 |
ENST00000542176.1 ENST00000278499.2 |
SESN3 |
sestrin 3 |
| chr3_+_57875711 | 0.72 |
ENST00000442599.2 |
SLMAP |
sarcolemma associated protein |
| chr11_-_119234876 | 0.72 |
ENST00000525735.1 |
USP2 |
ubiquitin specific peptidase 2 |
| chr17_-_39646116 | 0.70 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr4_+_146403912 | 0.69 |
ENST00000507367.1 ENST00000394092.2 ENST00000515385.1 |
SMAD1 |
SMAD family member 1 |
| chr3_+_111260856 | 0.69 |
ENST00000352690.4 |
CD96 |
CD96 molecule |
| chr7_-_25268104 | 0.68 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chr3_-_141747950 | 0.67 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_+_27665232 | 0.67 |
ENST00000543753.1 ENST00000288873.3 |
KRTCAP3 |
keratinocyte associated protein 3 |
| chr17_+_55163075 | 0.65 |
ENST00000571629.1 ENST00000570423.1 ENST00000575186.1 ENST00000573085.1 ENST00000572814.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr4_+_20255123 | 0.65 |
ENST00000504154.1 ENST00000273739.5 |
SLIT2 |
slit homolog 2 (Drosophila) |
| chr2_+_27665289 | 0.64 |
ENST00000407293.1 |
KRTCAP3 |
keratinocyte associated protein 3 |
| chr3_+_111260954 | 0.64 |
ENST00000283285.5 |
CD96 |
CD96 molecule |
| chr11_-_34535297 | 0.63 |
ENST00000532417.1 |
ELF5 |
E74-like factor 5 (ets domain transcription factor) |
| chr17_-_41623691 | 0.63 |
ENST00000545954.1 |
ETV4 |
ets variant 4 |
| chr4_+_95972822 | 0.62 |
ENST00000509540.1 ENST00000440890.2 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr6_+_13925318 | 0.62 |
ENST00000423553.2 ENST00000537388.1 |
RNF182 |
ring finger protein 182 |
| chr11_+_120039685 | 0.62 |
ENST00000530303.1 ENST00000319763.1 |
AP000679.2 |
Uncharacterized protein |
| chr10_+_71561649 | 0.61 |
ENST00000398978.3 ENST00000354547.3 ENST00000357811.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chrX_-_128788914 | 0.61 |
ENST00000429967.1 ENST00000307484.6 |
APLN |
apelin |
| chr1_+_84630367 | 0.60 |
ENST00000370680.1 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chrX_+_56259316 | 0.59 |
ENST00000468660.1 |
KLF8 |
Kruppel-like factor 8 |
| chr7_+_73245193 | 0.59 |
ENST00000340958.2 |
CLDN4 |
claudin 4 |
| chr7_-_16505440 | 0.59 |
ENST00000307068.4 |
SOSTDC1 |
sclerostin domain containing 1 |
| chr10_+_71561630 | 0.59 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr3_+_186648307 | 0.58 |
ENST00000457772.2 ENST00000455441.1 ENST00000427315.1 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr3_+_186648274 | 0.57 |
ENST00000169298.3 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr19_-_49864746 | 0.57 |
ENST00000598810.1 |
TEAD2 |
TEA domain family member 2 |
| chr17_-_41623716 | 0.57 |
ENST00000319349.5 |
ETV4 |
ets variant 4 |
| chr7_-_25702669 | 0.56 |
ENST00000446840.1 |
AC003090.1 |
AC003090.1 |
| chr3_+_57875738 | 0.56 |
ENST00000417128.1 ENST00000438794.1 |
SLMAP |
sarcolemma associated protein |
| chr2_+_162272605 | 0.56 |
ENST00000389554.3 |
TBR1 |
T-box, brain, 1 |
| chr9_+_116638562 | 0.55 |
ENST00000374126.5 ENST00000288466.7 |
ZNF618 |
zinc finger protein 618 |
| chr9_+_504674 | 0.55 |
ENST00000382297.2 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr10_+_24497704 | 0.53 |
ENST00000376456.4 ENST00000458595.1 |
KIAA1217 |
KIAA1217 |
| chr1_+_100111479 | 0.53 |
ENST00000263174.4 |
PALMD |
palmdelphin |
| chr10_+_24498060 | 0.53 |
ENST00000376454.3 ENST00000376452.3 |
KIAA1217 |
KIAA1217 |
| chr18_-_24237339 | 0.53 |
ENST00000580191.1 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
| chrX_-_24665208 | 0.53 |
ENST00000356768.4 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr8_-_139926236 | 0.53 |
ENST00000303045.6 ENST00000435777.1 |
COL22A1 |
collagen, type XXII, alpha 1 |
| chr5_+_140235469 | 0.52 |
ENST00000506939.2 ENST00000307360.5 |
PCDHA10 |
protocadherin alpha 10 |
| chr3_-_33700933 | 0.52 |
ENST00000480013.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr4_+_113152881 | 0.52 |
ENST00000274000.5 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr2_-_72375167 | 0.51 |
ENST00000001146.2 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr2_+_233734994 | 0.51 |
ENST00000331342.2 |
C2orf82 |
chromosome 2 open reading frame 82 |
| chr13_+_97928395 | 0.50 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chrX_+_37545012 | 0.50 |
ENST00000378616.3 |
XK |
X-linked Kx blood group (McLeod syndrome) |
| chr4_+_96012614 | 0.50 |
ENST00000264568.4 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr4_+_71494461 | 0.50 |
ENST00000396073.3 |
ENAM |
enamelin |
| chr5_+_140207536 | 0.50 |
ENST00000529310.1 ENST00000527624.1 |
PCDHA6 |
protocadherin alpha 6 |
| chr5_-_16936340 | 0.50 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr1_-_12677714 | 0.49 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chr2_-_74780176 | 0.49 |
ENST00000409549.1 |
LOXL3 |
lysyl oxidase-like 3 |
| chr7_+_39017504 | 0.48 |
ENST00000403058.1 |
POU6F2 |
POU class 6 homeobox 2 |
| chr11_-_34535332 | 0.47 |
ENST00000257832.2 ENST00000429939.2 |
ELF5 |
E74-like factor 5 (ets domain transcription factor) |
| chr14_-_65409502 | 0.47 |
ENST00000389614.5 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr16_-_4323015 | 0.46 |
ENST00000204517.6 |
TFAP4 |
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr4_-_123542224 | 0.46 |
ENST00000264497.3 |
IL21 |
interleukin 21 |
| chr12_-_48963829 | 0.46 |
ENST00000301046.2 ENST00000549817.1 |
LALBA |
lactalbumin, alpha- |
| chr8_-_145642267 | 0.45 |
ENST00000301305.3 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr15_-_52043722 | 0.45 |
ENST00000454181.2 |
LYSMD2 |
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr6_+_4776580 | 0.45 |
ENST00000397588.3 |
CDYL |
chromodomain protein, Y-like |
| chr11_-_75917569 | 0.44 |
ENST00000322563.3 |
WNT11 |
wingless-type MMTV integration site family, member 11 |
| chr4_+_71458012 | 0.43 |
ENST00000449493.2 |
AMBN |
ameloblastin (enamel matrix protein) |
| chr11_-_117747434 | 0.43 |
ENST00000529335.2 ENST00000530956.1 ENST00000260282.4 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr1_+_144339738 | 0.42 |
ENST00000538264.1 |
AL592284.1 |
Protein LOC642441 |
| chr1_-_54303949 | 0.42 |
ENST00000234725.8 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr2_-_118943930 | 0.42 |
ENST00000449075.1 ENST00000414886.1 ENST00000449819.1 |
AC093901.1 |
AC093901.1 |
| chr2_-_9143786 | 0.42 |
ENST00000462696.1 ENST00000305997.3 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
| chr1_-_216896780 | 0.42 |
ENST00000459955.1 ENST00000366937.1 ENST00000408911.3 ENST00000391890.3 |
ESRRG |
estrogen-related receptor gamma |
| chr10_+_134210672 | 0.42 |
ENST00000305233.5 ENST00000368609.4 |
PWWP2B |
PWWP domain containing 2B |
| chr4_+_30721968 | 0.42 |
ENST00000361762.2 |
PCDH7 |
protocadherin 7 |
| chr5_+_161494521 | 0.41 |
ENST00000356592.3 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chrX_-_24665353 | 0.40 |
ENST00000379144.2 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr16_-_4987065 | 0.40 |
ENST00000590782.2 ENST00000345988.2 |
PPL |
periplakin |
| chr18_+_28956740 | 0.40 |
ENST00000308128.4 ENST00000359747.4 |
DSG4 |
desmoglein 4 |
| chr10_-_61900762 | 0.40 |
ENST00000355288.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr17_-_27503770 | 0.40 |
ENST00000533112.1 |
MYO18A |
myosin XVIIIA |
| chr12_+_70760056 | 0.40 |
ENST00000258111.4 |
KCNMB4 |
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr20_+_20348740 | 0.40 |
ENST00000310227.1 |
INSM1 |
insulinoma-associated 1 |
| chr8_-_20161466 | 0.40 |
ENST00000381569.1 |
LZTS1 |
leucine zipper, putative tumor suppressor 1 |
| chr6_+_135502466 | 0.40 |
ENST00000367814.4 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr3_+_132757215 | 0.39 |
ENST00000321871.6 ENST00000393130.3 ENST00000514894.1 ENST00000512662.1 |
TMEM108 |
transmembrane protein 108 |
| chr4_+_113152978 | 0.38 |
ENST00000309703.6 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr17_-_41738931 | 0.38 |
ENST00000329168.3 ENST00000549132.1 |
MEOX1 |
mesenchyme homeobox 1 |
| chr9_+_84304628 | 0.38 |
ENST00000437181.1 |
RP11-154D17.1 |
RP11-154D17.1 |
| chr17_+_57297807 | 0.38 |
ENST00000284116.4 ENST00000581140.1 ENST00000581276.1 |
GDPD1 |
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr4_-_139163491 | 0.37 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr16_-_30122717 | 0.37 |
ENST00000566613.1 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr18_-_52989525 | 0.37 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr2_-_74781061 | 0.37 |
ENST00000264094.3 ENST00000393937.2 ENST00000409986.1 |
LOXL3 |
lysyl oxidase-like 3 |
| chr15_+_50474385 | 0.37 |
ENST00000267842.5 |
SLC27A2 |
solute carrier family 27 (fatty acid transporter), member 2 |
| chr1_-_54303934 | 0.37 |
ENST00000537333.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr1_+_196743912 | 0.37 |
ENST00000367425.4 |
CFHR3 |
complement factor H-related 3 |
| chr11_-_47207390 | 0.37 |
ENST00000539589.1 ENST00000528462.1 |
PACSIN3 |
protein kinase C and casein kinase substrate in neurons 3 |
| chr5_-_115872142 | 0.37 |
ENST00000510263.1 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr11_-_75017734 | 0.37 |
ENST00000532525.1 |
ARRB1 |
arrestin, beta 1 |
| chr15_-_65067773 | 0.37 |
ENST00000300069.4 |
RBPMS2 |
RNA binding protein with multiple splicing 2 |
| chr6_+_135502408 | 0.36 |
ENST00000341911.5 ENST00000442647.2 ENST00000316528.8 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr4_-_23891693 | 0.36 |
ENST00000264867.2 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr6_-_25874440 | 0.36 |
ENST00000361703.6 ENST00000397060.4 |
SLC17A3 |
solute carrier family 17 (organic anion transporter), member 3 |
| chr17_-_41739283 | 0.36 |
ENST00000393661.2 ENST00000318579.4 |
MEOX1 |
mesenchyme homeobox 1 |
| chr1_+_32739733 | 0.36 |
ENST00000333070.4 |
LCK |
lymphocyte-specific protein tyrosine kinase |
| chr18_-_53069419 | 0.35 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr1_+_196743943 | 0.35 |
ENST00000471440.2 ENST00000391985.3 |
CFHR3 |
complement factor H-related 3 |
| chr8_-_33424636 | 0.34 |
ENST00000256257.1 |
RNF122 |
ring finger protein 122 |
| chr4_-_123377880 | 0.34 |
ENST00000226730.4 |
IL2 |
interleukin 2 |
| chr1_-_53793584 | 0.34 |
ENST00000354412.3 ENST00000347547.2 ENST00000306052.6 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr13_-_99630233 | 0.34 |
ENST00000376460.1 ENST00000442173.1 |
DOCK9 |
dedicator of cytokinesis 9 |
| chr2_+_26568965 | 0.34 |
ENST00000260585.7 ENST00000447170.1 |
EPT1 |
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr10_+_71561704 | 0.34 |
ENST00000520267.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr3_-_141747439 | 0.34 |
ENST00000467667.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_-_127840021 | 0.34 |
ENST00000465909.2 |
SOGA3 |
SOGA family member 3 |
| chr1_+_27668505 | 0.34 |
ENST00000318074.5 |
SYTL1 |
synaptotagmin-like 1 |
| chr3_-_33686743 | 0.34 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr7_-_19184929 | 0.33 |
ENST00000275461.3 |
FERD3L |
Fer3-like bHLH transcription factor |
| chr3_+_171561127 | 0.33 |
ENST00000334567.5 ENST00000450693.1 |
TMEM212 |
transmembrane protein 212 |
| chr17_-_7297833 | 0.33 |
ENST00000571802.1 ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr1_+_93544791 | 0.33 |
ENST00000545708.1 ENST00000540243.1 ENST00000370298.4 |
MTF2 |
metal response element binding transcription factor 2 |
| chr3_+_178253993 | 0.32 |
ENST00000420517.2 ENST00000452583.1 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr1_-_155948318 | 0.32 |
ENST00000361247.4 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr14_-_23451845 | 0.32 |
ENST00000262713.2 |
AJUBA |
ajuba LIM protein |
| chr6_+_135502501 | 0.32 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr11_-_104769141 | 0.32 |
ENST00000508062.1 ENST00000422698.2 |
CASP12 |
caspase 12 (gene/pseudogene) |
| chr5_-_76935513 | 0.32 |
ENST00000306422.3 |
OTP |
orthopedia homeobox |
| chr4_-_85419603 | 0.32 |
ENST00000295886.4 |
NKX6-1 |
NK6 homeobox 1 |
| chr6_-_127840336 | 0.31 |
ENST00000525778.1 |
SOGA3 |
SOGA family member 3 |
| chr10_+_123923205 | 0.31 |
ENST00000369004.3 ENST00000260733.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr6_+_138483058 | 0.31 |
ENST00000251691.4 |
KIAA1244 |
KIAA1244 |
| chr3_+_52813932 | 0.31 |
ENST00000537050.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr20_-_23066953 | 0.31 |
ENST00000246006.4 |
CD93 |
CD93 molecule |
| chr4_-_72649763 | 0.31 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr1_-_226496898 | 0.31 |
ENST00000481685.1 |
LIN9 |
lin-9 homolog (C. elegans) |
| chr14_-_57272366 | 0.31 |
ENST00000554788.1 ENST00000554845.1 ENST00000408990.3 |
OTX2 |
orthodenticle homeobox 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0032771 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.6 | 1.8 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.6 | 2.3 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.5 | 1.6 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.5 | 1.5 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.5 | 1.4 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.4 | 4.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.4 | 1.6 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.4 | 3.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.4 | 1.1 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.4 | 1.1 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.3 | 0.9 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.3 | 0.9 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.3 | 0.8 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.2 | 0.2 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.2 | 1.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 0.6 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.2 | 1.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 0.6 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 | 0.5 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 1.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 0.8 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 1.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.2 | 0.5 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 1.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.2 | 0.5 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.2 | 0.3 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.4 | GO:0060775 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.1 | 0.7 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 1.6 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.6 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 0.4 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 | 0.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.4 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.4 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.7 | GO:0061056 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.1 | 0.5 | GO:0048867 | ganglion mother cell fate determination(GO:0007402) stem cell fate determination(GO:0048867) |
| 0.1 | 1.3 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 0.5 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.1 | 0.3 | GO:0021913 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.1 | 0.6 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.3 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.1 | 0.6 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.4 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.7 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.4 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.6 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 0.3 | GO:0060061 | Spemann organizer formation(GO:0060061) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.1 | 1.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.5 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) |
| 0.1 | 1.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 1.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.9 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 2.0 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 2.1 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 0.4 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.3 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 | 0.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.2 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.5 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.1 | 0.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.6 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 1.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.7 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.3 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.1 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.3 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.1 | 0.5 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.3 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 1.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.3 | GO:0000436 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.0 | 0.2 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.1 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.4 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.6 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.4 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 1.9 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 5.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.5 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.8 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0021592 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) |
| 0.0 | 0.4 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.3 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 1.4 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.0 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) |
| 0.0 | 0.3 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.3 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 1.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.3 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.0 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.0 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) regulation of response to macrophage colony-stimulating factor(GO:1903969) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.0 | 0.1 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.6 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.0 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 1.0 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.3 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.4 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.4 | GO:0006629 | lipid metabolic process(GO:0006629) |
| 0.0 | 0.4 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.4 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.0 | 0.3 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0021612 | facial nerve structural organization(GO:0021612) |
| 0.0 | 1.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0030879 | mammary gland development(GO:0030879) |
| 0.0 | 0.2 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:0035995 | skeletal muscle myosin thick filament assembly(GO:0030241) detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0061113 | elastin metabolic process(GO:0051541) pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 1.0 | 2.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.6 | 1.8 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 1.8 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.4 | 1.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.3 | 1.3 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 1.6 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 0.6 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 4.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.6 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.8 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.0 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 1.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 2.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 2.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 2.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 1.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 2.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 4.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 2.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.3 | 2.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 1.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 2.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 2.0 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.5 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 1.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 1.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.4 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.6 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 2.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.6 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 0.2 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.1 | 0.4 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 1.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.3 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.2 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.8 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 1.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 2.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.5 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.6 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.6 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 2.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 3.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.4 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0015245 | fatty acid transporter activity(GO:0015245) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 2.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 4.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 6.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 2.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 2.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 5.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 6.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 2.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |