Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for LHX6
Z-value: 0.16
Transcription factors associated with LHX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX6
|
ENSG00000106852.11 | LHX6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX6 | hg19_v2_chr9_-_124991124_124991246, hg19_v2_chr9_-_124989804_124989865 | -0.25 | 5.5e-01 | Click! |
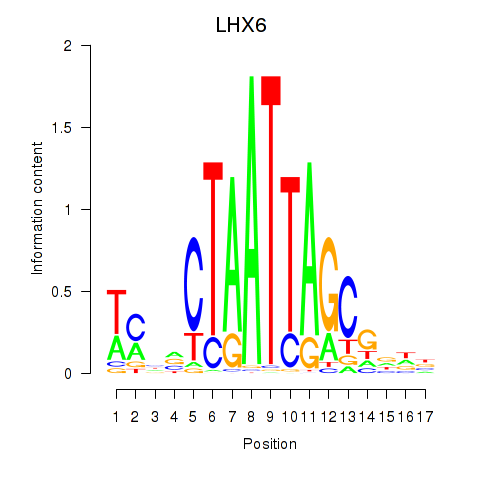
Activity profile of LHX6 motif
Sorted Z-values of LHX6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91546926 | 0.32 |
ENST00000550758.1 |
DCN |
decorin |
| chr4_-_138453606 | 0.30 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr11_+_5710919 | 0.30 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr1_-_236228403 | 0.28 |
ENST00000366595.3 |
NID1 |
nidogen 1 |
| chr6_-_167040731 | 0.28 |
ENST00000265678.4 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr1_-_236228417 | 0.27 |
ENST00000264187.6 |
NID1 |
nidogen 1 |
| chr5_-_20575959 | 0.23 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr4_-_138453559 | 0.21 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr10_-_28571015 | 0.20 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr2_+_220143989 | 0.19 |
ENST00000336576.5 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr1_+_145438469 | 0.19 |
ENST00000369317.4 |
TXNIP |
thioredoxin interacting protein |
| chr2_+_220144052 | 0.18 |
ENST00000425450.1 ENST00000392086.4 ENST00000421532.1 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr16_+_55512742 | 0.16 |
ENST00000568715.1 ENST00000219070.4 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr1_+_151735431 | 0.14 |
ENST00000321531.5 ENST00000315067.8 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr7_+_149571045 | 0.13 |
ENST00000479613.1 ENST00000606024.1 ENST00000464662.1 ENST00000425642.2 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
| chr6_+_122931366 | 0.12 |
ENST00000368452.2 ENST00000368448.1 ENST00000392490.1 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr12_+_7014064 | 0.11 |
ENST00000443597.2 |
LRRC23 |
leucine rich repeat containing 23 |
| chr12_+_7013897 | 0.11 |
ENST00000007969.8 ENST00000323702.5 |
LRRC23 |
leucine rich repeat containing 23 |
| chr3_-_99569821 | 0.11 |
ENST00000487087.1 |
FILIP1L |
filamin A interacting protein 1-like |
| chr14_+_74034310 | 0.11 |
ENST00000538782.1 |
ACOT2 |
acyl-CoA thioesterase 2 |
| chr8_-_7243080 | 0.10 |
ENST00000400156.4 |
ZNF705G |
zinc finger protein 705G |
| chr14_-_52535712 | 0.10 |
ENST00000216286.5 ENST00000541773.1 |
NID2 |
nidogen 2 (osteonidogen) |
| chrX_-_47509887 | 0.10 |
ENST00000247161.3 ENST00000592066.1 ENST00000376983.3 |
ELK1 |
ELK1, member of ETS oncogene family |
| chr19_-_40931891 | 0.10 |
ENST00000357949.4 |
SERTAD1 |
SERTA domain containing 1 |
| chr16_-_66583701 | 0.10 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr22_-_39190116 | 0.10 |
ENST00000406622.1 ENST00000216068.4 ENST00000406199.3 |
SUN2 DNAL4 |
Sad1 and UNC84 domain containing 2 dynein, axonemal, light chain 4 |
| chr17_+_57408994 | 0.10 |
ENST00000312655.4 |
YPEL2 |
yippee-like 2 (Drosophila) |
| chr11_+_77532155 | 0.10 |
ENST00000532481.1 ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr1_-_48866517 | 0.09 |
ENST00000371841.1 |
SPATA6 |
spermatogenesis associated 6 |
| chr2_-_3521518 | 0.09 |
ENST00000382093.5 |
ADI1 |
acireductone dioxygenase 1 |
| chr11_+_77532233 | 0.09 |
ENST00000525409.1 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr3_-_105588231 | 0.09 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr5_+_162864575 | 0.09 |
ENST00000512163.1 ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1 |
cyclin G1 |
| chr9_+_12775011 | 0.09 |
ENST00000319264.3 |
LURAP1L |
leucine rich adaptor protein 1-like |
| chr8_-_7220490 | 0.09 |
ENST00000400078.2 |
ZNF705G |
zinc finger protein 705G |
| chr12_-_5352315 | 0.09 |
ENST00000536518.1 |
RP11-319E16.1 |
RP11-319E16.1 |
| chr17_-_40337470 | 0.09 |
ENST00000293330.1 |
HCRT |
hypocretin (orexin) neuropeptide precursor |
| chr5_-_147286065 | 0.09 |
ENST00000318315.4 ENST00000515291.1 |
C5orf46 |
chromosome 5 open reading frame 46 |
| chr17_+_66511540 | 0.08 |
ENST00000588188.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr7_-_14029515 | 0.08 |
ENST00000430479.1 ENST00000405218.2 ENST00000343495.5 |
ETV1 |
ets variant 1 |
| chr1_-_232598163 | 0.08 |
ENST00000308942.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr2_+_166095898 | 0.08 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr8_-_13134045 | 0.08 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr10_+_13142225 | 0.08 |
ENST00000378747.3 |
OPTN |
optineurin |
| chr17_+_59489112 | 0.08 |
ENST00000335108.2 |
C17orf82 |
chromosome 17 open reading frame 82 |
| chr3_-_105587879 | 0.08 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr7_+_100209725 | 0.07 |
ENST00000223054.4 |
MOSPD3 |
motile sperm domain containing 3 |
| chr7_+_100209979 | 0.07 |
ENST00000493970.1 ENST00000379527.2 |
MOSPD3 |
motile sperm domain containing 3 |
| chr2_-_152118352 | 0.07 |
ENST00000331426.5 |
RBM43 |
RNA binding motif protein 43 |
| chr18_-_65184217 | 0.07 |
ENST00000310045.7 |
DSEL |
dermatan sulfate epimerase-like |
| chr1_-_36906474 | 0.07 |
ENST00000433045.2 |
OSCP1 |
organic solute carrier partner 1 |
| chr7_+_100210133 | 0.07 |
ENST00000393950.2 ENST00000424091.2 |
MOSPD3 |
motile sperm domain containing 3 |
| chr12_-_102455846 | 0.07 |
ENST00000545679.1 |
CCDC53 |
coiled-coil domain containing 53 |
| chr13_-_76111945 | 0.07 |
ENST00000355801.4 ENST00000406936.3 |
COMMD6 |
COMM domain containing 6 |
| chr19_-_12833361 | 0.07 |
ENST00000592287.1 |
TNPO2 |
transportin 2 |
| chr22_+_24309089 | 0.07 |
ENST00000215770.5 |
DDTL |
D-dopachrome tautomerase-like |
| chr14_+_100842735 | 0.06 |
ENST00000554998.1 ENST00000402312.3 ENST00000335290.6 ENST00000554175.1 |
WDR25 |
WD repeat domain 25 |
| chr12_-_56753858 | 0.06 |
ENST00000314128.4 ENST00000557235.1 ENST00000418572.2 |
STAT2 |
signal transducer and activator of transcription 2, 113kDa |
| chr12_-_102455902 | 0.06 |
ENST00000240079.6 |
CCDC53 |
coiled-coil domain containing 53 |
| chr16_-_66584059 | 0.06 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr19_+_12902289 | 0.06 |
ENST00000302754.4 |
JUNB |
jun B proto-oncogene |
| chr22_+_17956618 | 0.06 |
ENST00000262608.8 |
CECR2 |
cat eye syndrome chromosome region, candidate 2 |
| chr9_+_104161123 | 0.06 |
ENST00000374861.3 ENST00000339664.2 ENST00000259395.4 |
ZNF189 |
zinc finger protein 189 |
| chr2_+_201936458 | 0.06 |
ENST00000237889.4 |
NDUFB3 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr6_+_148663729 | 0.06 |
ENST00000367467.3 |
SASH1 |
SAM and SH3 domain containing 1 |
| chr15_+_40697988 | 0.06 |
ENST00000487418.2 ENST00000479013.2 |
IVD |
isovaleryl-CoA dehydrogenase |
| chr11_-_76155700 | 0.06 |
ENST00000572035.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chr11_+_33061543 | 0.06 |
ENST00000432887.1 ENST00000528898.1 ENST00000531632.2 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
| chr9_+_80912059 | 0.06 |
ENST00000347159.2 ENST00000376588.3 |
PSAT1 |
phosphoserine aminotransferase 1 |
| chr8_-_42623747 | 0.06 |
ENST00000534622.1 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr19_-_10121144 | 0.06 |
ENST00000264828.3 |
COL5A3 |
collagen, type V, alpha 3 |
| chr11_+_67798363 | 0.05 |
ENST00000525628.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr6_-_85474219 | 0.05 |
ENST00000369663.5 |
TBX18 |
T-box 18 |
| chr6_+_26402517 | 0.05 |
ENST00000414912.2 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chr14_+_104182061 | 0.05 |
ENST00000216602.6 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr6_+_46661575 | 0.05 |
ENST00000450697.1 |
TDRD6 |
tudor domain containing 6 |
| chr14_-_99947121 | 0.05 |
ENST00000329331.3 ENST00000436070.2 |
SETD3 |
SET domain containing 3 |
| chr11_+_67798114 | 0.05 |
ENST00000453471.2 ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr17_+_18086392 | 0.05 |
ENST00000541285.1 |
ALKBH5 |
alkB, alkylation repair homolog 5 (E. coli) |
| chr10_+_13141441 | 0.05 |
ENST00000263036.5 |
OPTN |
optineurin |
| chr14_+_104182105 | 0.05 |
ENST00000311141.2 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr17_-_79623597 | 0.05 |
ENST00000574024.1 ENST00000331056.5 |
PDE6G |
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr17_+_74261413 | 0.05 |
ENST00000587913.1 |
UBALD2 |
UBA-like domain containing 2 |
| chr7_+_120628731 | 0.05 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr21_-_38639813 | 0.05 |
ENST00000309117.6 ENST00000398998.1 |
DSCR3 |
Down syndrome critical region gene 3 |
| chr21_-_22175450 | 0.05 |
ENST00000435279.2 |
LINC00320 |
long intergenic non-protein coding RNA 320 |
| chr1_-_173886491 | 0.05 |
ENST00000367698.3 |
SERPINC1 |
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr11_+_67798090 | 0.05 |
ENST00000313468.5 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr5_-_54830784 | 0.05 |
ENST00000264775.5 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
| chr11_-_76155618 | 0.05 |
ENST00000530759.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chr11_-_124981475 | 0.05 |
ENST00000532156.1 ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218 |
transmembrane protein 218 |
| chr12_-_49418407 | 0.05 |
ENST00000526209.1 |
KMT2D |
lysine (K)-specific methyltransferase 2D |
| chr10_+_13141585 | 0.05 |
ENST00000378764.2 |
OPTN |
optineurin |
| chr18_+_44526786 | 0.05 |
ENST00000245121.5 ENST00000356157.7 |
KATNAL2 |
katanin p60 subunit A-like 2 |
| chr7_-_86849883 | 0.05 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr5_-_54830871 | 0.05 |
ENST00000307259.8 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
| chr18_-_5238525 | 0.05 |
ENST00000581170.1 ENST00000579933.1 ENST00000581067.1 |
RP11-835E18.5 LINC00526 |
RP11-835E18.5 long intergenic non-protein coding RNA 526 |
| chr17_-_1532106 | 0.04 |
ENST00000301335.5 ENST00000382147.4 |
SLC43A2 |
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr6_+_39760129 | 0.04 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr21_-_22175341 | 0.04 |
ENST00000416768.1 ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320 |
long intergenic non-protein coding RNA 320 |
| chr18_+_5238549 | 0.04 |
ENST00000580684.1 |
LINC00667 |
long intergenic non-protein coding RNA 667 |
| chr12_-_2113583 | 0.04 |
ENST00000397173.4 ENST00000280665.6 |
DCP1B |
decapping mRNA 1B |
| chr6_+_26402465 | 0.04 |
ENST00000476549.2 ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chrX_+_77154935 | 0.04 |
ENST00000481445.1 |
COX7B |
cytochrome c oxidase subunit VIIb |
| chr17_-_40828969 | 0.04 |
ENST00000591022.1 ENST00000587627.1 ENST00000293349.6 |
PLEKHH3 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr11_+_76156045 | 0.04 |
ENST00000533988.1 ENST00000524490.1 ENST00000334736.3 ENST00000343878.3 ENST00000533972.1 |
C11orf30 |
chromosome 11 open reading frame 30 |
| chr16_+_31724552 | 0.04 |
ENST00000539915.1 ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720 |
zinc finger protein 720 |
| chr12_+_8666126 | 0.04 |
ENST00000299665.2 |
CLEC4D |
C-type lectin domain family 4, member D |
| chr10_-_99447024 | 0.04 |
ENST00000370626.3 |
AVPI1 |
arginine vasopressin-induced 1 |
| chr11_-_124180733 | 0.04 |
ENST00000357821.2 |
OR8D1 |
olfactory receptor, family 8, subfamily D, member 1 |
| chr6_-_33385902 | 0.04 |
ENST00000374500.5 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr6_-_33385823 | 0.04 |
ENST00000494751.1 ENST00000374496.3 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr5_-_171881362 | 0.04 |
ENST00000519643.1 |
SH3PXD2B |
SH3 and PX domains 2B |
| chr4_-_103749205 | 0.04 |
ENST00000508249.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_33385870 | 0.04 |
ENST00000488034.1 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr4_-_103748880 | 0.04 |
ENST00000453744.2 ENST00000349311.8 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr13_+_98086445 | 0.03 |
ENST00000245304.4 |
RAP2A |
RAP2A, member of RAS oncogene family |
| chr6_-_33385854 | 0.03 |
ENST00000488478.1 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr1_-_1709845 | 0.03 |
ENST00000341426.5 ENST00000344463.4 |
NADK |
NAD kinase |
| chr3_-_157221380 | 0.03 |
ENST00000468233.1 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
| chr6_+_36165133 | 0.03 |
ENST00000446974.1 ENST00000454960.1 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
| chr6_-_33385655 | 0.03 |
ENST00000440279.3 ENST00000607266.1 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr7_-_72971934 | 0.03 |
ENST00000411832.1 |
BCL7B |
B-cell CLL/lymphoma 7B |
| chr1_-_1710229 | 0.03 |
ENST00000341991.3 |
NADK |
NAD kinase |
| chr17_+_41323204 | 0.03 |
ENST00000542611.1 ENST00000590996.1 ENST00000389312.4 ENST00000589872.1 |
NBR1 |
neighbor of BRCA1 gene 1 |
| chr7_-_140714739 | 0.03 |
ENST00000467334.1 ENST00000324787.5 |
MRPS33 |
mitochondrial ribosomal protein S33 |
| chr2_+_65454926 | 0.03 |
ENST00000542850.1 ENST00000377982.4 |
ACTR2 |
ARP2 actin-related protein 2 homolog (yeast) |
| chr7_+_70597109 | 0.03 |
ENST00000333538.5 |
WBSCR17 |
Williams-Beuren syndrome chromosome region 17 |
| chr18_+_616672 | 0.03 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr12_+_7014126 | 0.03 |
ENST00000415834.1 ENST00000436789.1 |
LRRC23 |
leucine rich repeat containing 23 |
| chr9_+_117904097 | 0.03 |
ENST00000374016.1 |
DEC1 |
deleted in esophageal cancer 1 |
| chr14_-_53258314 | 0.03 |
ENST00000216410.3 ENST00000557604.1 |
GNPNAT1 |
glucosamine-phosphate N-acetyltransferase 1 |
| chr14_+_57671888 | 0.03 |
ENST00000391612.1 |
AL391152.1 |
AL391152.1 |
| chr10_-_72142345 | 0.03 |
ENST00000373224.1 ENST00000446961.1 ENST00000358141.2 ENST00000357631.2 |
LRRC20 |
leucine rich repeat containing 20 |
| chr3_-_157221357 | 0.03 |
ENST00000494677.1 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
| chr15_-_77712477 | 0.03 |
ENST00000560626.2 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
| chr3_+_108541608 | 0.03 |
ENST00000426646.1 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr20_+_5731083 | 0.03 |
ENST00000445603.1 ENST00000442185.1 |
C20orf196 |
chromosome 20 open reading frame 196 |
| chr14_+_52456327 | 0.03 |
ENST00000556760.1 |
C14orf166 |
chromosome 14 open reading frame 166 |
| chr20_-_35807741 | 0.03 |
ENST00000434295.1 ENST00000441008.2 ENST00000400441.3 ENST00000343811.4 |
MROH8 |
maestro heat-like repeat family member 8 |
| chr1_+_241695424 | 0.03 |
ENST00000366558.3 ENST00000366559.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr19_+_19303008 | 0.03 |
ENST00000353145.1 ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK |
regulatory factor X-associated ankyrin-containing protein |
| chr14_+_76127529 | 0.03 |
ENST00000556977.1 ENST00000557636.1 ENST00000286650.5 ENST00000298832.9 |
TTLL5 |
tubulin tyrosine ligase-like family, member 5 |
| chr12_+_118573663 | 0.03 |
ENST00000261313.2 |
PEBP1 |
phosphatidylethanolamine binding protein 1 |
| chr16_+_15744078 | 0.03 |
ENST00000396354.1 ENST00000570727.1 |
NDE1 |
nudE neurodevelopment protein 1 |
| chr2_+_242089833 | 0.03 |
ENST00000404405.3 ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7 |
protein phosphatase 1, regulatory subunit 7 |
| chr6_-_33285505 | 0.03 |
ENST00000431845.2 |
ZBTB22 |
zinc finger and BTB domain containing 22 |
| chr10_-_48416849 | 0.03 |
ENST00000249598.1 |
GDF2 |
growth differentiation factor 2 |
| chr1_-_13390765 | 0.03 |
ENST00000357367.2 |
PRAMEF8 |
PRAME family member 8 |
| chr2_+_65454863 | 0.03 |
ENST00000260641.5 |
ACTR2 |
ARP2 actin-related protein 2 homolog (yeast) |
| chr1_+_152943122 | 0.03 |
ENST00000328051.2 |
SPRR4 |
small proline-rich protein 4 |
| chr10_-_99205607 | 0.03 |
ENST00000477692.2 ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1 |
exosome component 1 |
| chr6_+_132455118 | 0.03 |
ENST00000458028.1 |
LINC01013 |
long intergenic non-protein coding RNA 1013 |
| chr11_-_236326 | 0.03 |
ENST00000525237.1 ENST00000532956.1 ENST00000525319.1 ENST00000524564.1 ENST00000382743.4 |
SIRT3 |
sirtuin 3 |
| chr17_-_73937116 | 0.03 |
ENST00000586717.1 ENST00000389570.4 ENST00000319129.5 |
FBF1 |
Fas (TNFRSF6) binding factor 1 |
| chr7_-_99277610 | 0.03 |
ENST00000343703.5 ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5 |
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr1_+_64669294 | 0.03 |
ENST00000371077.5 |
UBE2U |
ubiquitin-conjugating enzyme E2U (putative) |
| chr12_+_21525818 | 0.03 |
ENST00000240652.3 ENST00000542023.1 ENST00000537593.1 |
IAPP |
islet amyloid polypeptide |
| chr3_+_108541545 | 0.03 |
ENST00000295756.6 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr18_+_5238055 | 0.02 |
ENST00000582363.1 ENST00000582008.1 ENST00000580082.1 |
LINC00667 |
long intergenic non-protein coding RNA 667 |
| chr19_-_19302931 | 0.02 |
ENST00000444486.3 ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B MEF2BNB MEF2B |
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr4_-_103749105 | 0.02 |
ENST00000394801.4 ENST00000394804.2 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr3_-_157221128 | 0.02 |
ENST00000392833.2 ENST00000362010.2 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
| chr16_+_6533729 | 0.02 |
ENST00000551752.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr6_-_30585009 | 0.02 |
ENST00000376511.2 |
PPP1R10 |
protein phosphatase 1, regulatory subunit 10 |
| chr5_-_162887054 | 0.02 |
ENST00000517501.1 |
NUDCD2 |
NudC domain containing 2 |
| chr13_+_40229764 | 0.02 |
ENST00000416691.1 ENST00000422759.2 ENST00000455146.3 |
COG6 |
component of oligomeric golgi complex 6 |
| chr2_-_192711968 | 0.02 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chr7_+_142458507 | 0.02 |
ENST00000492062.1 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
| chr5_-_162887071 | 0.02 |
ENST00000302764.4 |
NUDCD2 |
NudC domain containing 2 |
| chr6_+_36164487 | 0.02 |
ENST00000357641.6 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
| chr3_+_156799587 | 0.02 |
ENST00000469196.1 |
RP11-6F2.5 |
RP11-6F2.5 |
| chr1_+_241695670 | 0.02 |
ENST00000366557.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr4_-_103749313 | 0.02 |
ENST00000394803.5 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr9_-_4666421 | 0.02 |
ENST00000381895.5 |
SPATA6L |
spermatogenesis associated 6-like |
| chr17_-_1531635 | 0.02 |
ENST00000571650.1 |
SLC43A2 |
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr17_-_4871085 | 0.02 |
ENST00000575142.1 ENST00000206020.3 |
SPAG7 |
sperm associated antigen 7 |
| chr11_-_18548426 | 0.02 |
ENST00000357193.3 ENST00000536719.1 |
TSG101 |
tumor susceptibility 101 |
| chr17_+_80416050 | 0.02 |
ENST00000579198.1 ENST00000390006.4 ENST00000580296.1 |
NARF |
nuclear prelamin A recognition factor |
| chr4_-_103749179 | 0.02 |
ENST00000502690.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr11_-_102714534 | 0.02 |
ENST00000299855.5 |
MMP3 |
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr17_-_40829026 | 0.02 |
ENST00000412503.1 |
PLEKHH3 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr2_-_74618964 | 0.02 |
ENST00000417090.1 ENST00000409868.1 |
DCTN1 |
dynactin 1 |
| chr3_+_40498783 | 0.02 |
ENST00000338970.6 ENST00000396203.2 ENST00000416518.1 |
RPL14 |
ribosomal protein L14 |
| chr13_-_81801115 | 0.02 |
ENST00000567258.1 |
LINC00564 |
long intergenic non-protein coding RNA 564 |
| chr13_-_41593425 | 0.02 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr16_+_12059050 | 0.02 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr19_+_41869894 | 0.02 |
ENST00000413014.2 |
TMEM91 |
transmembrane protein 91 |
| chr16_+_29817399 | 0.02 |
ENST00000545521.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr5_-_159846399 | 0.02 |
ENST00000297151.4 |
SLU7 |
SLU7 splicing factor homolog (S. cerevisiae) |
| chr1_-_157789850 | 0.02 |
ENST00000491942.1 ENST00000358292.3 ENST00000368176.3 |
FCRL1 |
Fc receptor-like 1 |
| chr16_+_6533380 | 0.02 |
ENST00000552089.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_+_201936707 | 0.02 |
ENST00000433898.1 ENST00000454214.1 |
NDUFB3 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr5_+_174151536 | 0.02 |
ENST00000239243.6 ENST00000507785.1 |
MSX2 |
msh homeobox 2 |
| chr17_+_41476327 | 0.02 |
ENST00000320033.4 |
ARL4D |
ADP-ribosylation factor-like 4D |
| chr18_-_44181442 | 0.01 |
ENST00000398722.4 |
LOXHD1 |
lipoxygenase homology domains 1 |
| chr11_-_33913708 | 0.01 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr9_-_127905736 | 0.01 |
ENST00000336505.6 ENST00000373549.4 |
SCAI |
suppressor of cancer cell invasion |
| chr2_+_11682790 | 0.01 |
ENST00000389825.3 ENST00000381483.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr3_+_67705121 | 0.01 |
ENST00000464420.1 ENST00000482677.1 |
RP11-81N13.1 |
RP11-81N13.1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.2 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.2 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |