Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for LMX1B_MNX1_RAX2
Z-value: 0.63
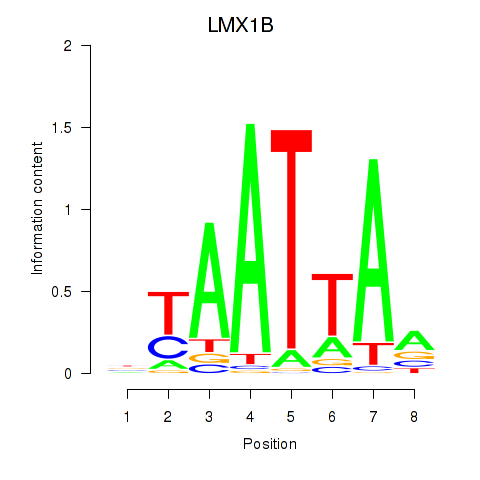
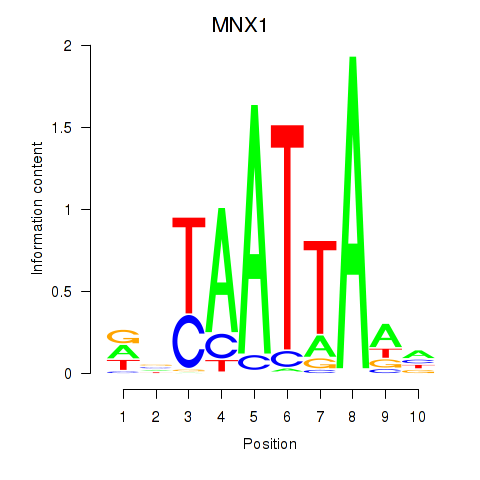
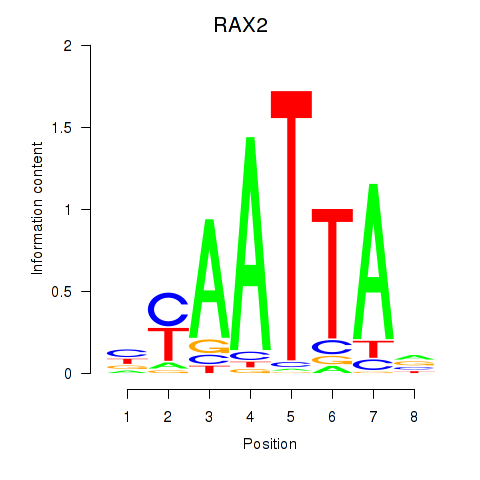
Transcription factors associated with LMX1B_MNX1_RAX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LMX1B
|
ENSG00000136944.13 | LMX1B |
|
MNX1
|
ENSG00000130675.10 | MNX1 |
|
RAX2
|
ENSG00000173976.11 | RAX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MNX1 | hg19_v2_chr7_-_156803329_156803362 | 0.46 | 2.6e-01 | Click! |
| LMX1B | hg19_v2_chr9_+_129376722_129376748 | 0.41 | 3.1e-01 | Click! |
| RAX2 | hg19_v2_chr19_-_3772209_3772236 | 0.30 | 4.8e-01 | Click! |
Activity profile of LMX1B_MNX1_RAX2 motif
Sorted Z-values of LMX1B_MNX1_RAX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of LMX1B_MNX1_RAX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_28123206 | 1.05 |
ENST00000542963.1 ENST00000535992.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr12_-_28122980 | 1.02 |
ENST00000395868.3 ENST00000534890.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr9_+_12693336 | 1.00 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr1_-_242612779 | 0.90 |
ENST00000427495.1 |
PLD5 |
phospholipase D family, member 5 |
| chr5_+_66300446 | 0.81 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_160370344 | 0.74 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr3_-_151034734 | 0.70 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr19_-_51522955 | 0.63 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr6_+_130339710 | 0.54 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr2_+_68961934 | 0.52 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr2_+_68961905 | 0.52 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr18_+_29027696 | 0.38 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr4_+_40198527 | 0.38 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr21_-_42219065 | 0.37 |
ENST00000400454.1 |
DSCAM |
Down syndrome cell adhesion molecule |
| chr8_-_42234745 | 0.37 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chr8_-_139926236 | 0.36 |
ENST00000303045.6 ENST00000435777.1 |
COL22A1 |
collagen, type XXII, alpha 1 |
| chr1_+_62439037 | 0.35 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr3_+_111718036 | 0.35 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr3_+_111717600 | 0.35 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr4_+_69313145 | 0.33 |
ENST00000305363.4 |
TMPRSS11E |
transmembrane protease, serine 11E |
| chr3_+_111718173 | 0.32 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr7_-_25268104 | 0.32 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chr4_-_25865159 | 0.31 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr3_+_111717511 | 0.31 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr18_+_34124507 | 0.31 |
ENST00000591635.1 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr8_+_105235572 | 0.30 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr5_-_24645078 | 0.30 |
ENST00000264463.4 |
CDH10 |
cadherin 10, type 2 (T2-cadherin) |
| chr11_-_124190184 | 0.29 |
ENST00000357438.2 |
OR8D2 |
olfactory receptor, family 8, subfamily D, member 2 |
| chr4_+_71458012 | 0.29 |
ENST00000449493.2 |
AMBN |
ameloblastin (enamel matrix protein) |
| chr2_+_68962014 | 0.29 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr12_+_41831485 | 0.28 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr19_-_51523412 | 0.28 |
ENST00000391805.1 ENST00000599077.1 |
KLK10 |
kallikrein-related peptidase 10 |
| chr4_-_74486217 | 0.26 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_-_136847099 | 0.26 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr19_-_51523275 | 0.26 |
ENST00000309958.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr13_-_46716969 | 0.25 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr12_+_4385230 | 0.25 |
ENST00000536537.1 |
CCND2 |
cyclin D2 |
| chr6_-_136788001 | 0.25 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chr2_-_136678123 | 0.25 |
ENST00000422708.1 |
DARS |
aspartyl-tRNA synthetase |
| chr6_-_136847610 | 0.24 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr12_+_107712173 | 0.24 |
ENST00000280758.5 ENST00000420571.2 |
BTBD11 |
BTB (POZ) domain containing 11 |
| chrX_+_105937068 | 0.23 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr8_-_109799793 | 0.23 |
ENST00000297459.3 |
TMEM74 |
transmembrane protein 74 |
| chr5_-_1882858 | 0.22 |
ENST00000511126.1 ENST00000231357.2 |
IRX4 |
iroquois homeobox 4 |
| chr20_-_50722183 | 0.22 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr10_+_98741041 | 0.22 |
ENST00000286067.2 |
C10orf12 |
chromosome 10 open reading frame 12 |
| chr3_-_27764190 | 0.21 |
ENST00000537516.1 |
EOMES |
eomesodermin |
| chr12_+_81110684 | 0.20 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr1_+_107683436 | 0.20 |
ENST00000370068.1 |
NTNG1 |
netrin G1 |
| chr15_-_68497657 | 0.20 |
ENST00000448060.2 ENST00000467889.1 |
CALML4 |
calmodulin-like 4 |
| chr17_-_64225508 | 0.19 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr13_-_86373536 | 0.19 |
ENST00000400286.2 |
SLITRK6 |
SLIT and NTRK-like family, member 6 |
| chr3_-_33686743 | 0.19 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr8_+_101170563 | 0.19 |
ENST00000520508.1 ENST00000388798.2 |
SPAG1 |
sperm associated antigen 1 |
| chr6_-_32157947 | 0.18 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr3_-_33686925 | 0.18 |
ENST00000485378.2 ENST00000313350.6 ENST00000487200.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr10_+_24497704 | 0.18 |
ENST00000376456.4 ENST00000458595.1 |
KIAA1217 |
KIAA1217 |
| chr3_+_130569429 | 0.17 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr17_-_9694614 | 0.17 |
ENST00000330255.5 ENST00000571134.1 |
DHRS7C |
dehydrogenase/reductase (SDR family) member 7C |
| chr4_-_139163491 | 0.17 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr3_+_15045419 | 0.16 |
ENST00000406272.2 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr7_-_92855762 | 0.16 |
ENST00000453812.2 ENST00000394468.2 |
HEPACAM2 |
HEPACAM family member 2 |
| chr1_-_234667504 | 0.16 |
ENST00000421207.1 ENST00000435574.1 |
RP5-855F14.1 |
RP5-855F14.1 |
| chr7_-_22234381 | 0.16 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_+_234826016 | 0.15 |
ENST00000324695.4 ENST00000433712.2 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
| chr14_-_78083112 | 0.15 |
ENST00000216484.2 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
| chr12_-_95510743 | 0.15 |
ENST00000551521.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr15_+_75080883 | 0.15 |
ENST00000567571.1 |
CSK |
c-src tyrosine kinase |
| chr4_-_77328458 | 0.14 |
ENST00000388914.3 ENST00000434846.2 |
CCDC158 |
coiled-coil domain containing 158 |
| chr6_-_161695074 | 0.14 |
ENST00000457520.2 ENST00000366906.5 ENST00000320285.4 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr12_+_83080659 | 0.14 |
ENST00000321196.3 |
TMTC2 |
transmembrane and tetratricopeptide repeat containing 2 |
| chr1_-_185597619 | 0.14 |
ENST00000608417.1 ENST00000436955.1 |
GS1-204I12.1 |
GS1-204I12.1 |
| chr17_-_39646116 | 0.13 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr15_+_76352178 | 0.13 |
ENST00000388942.3 |
C15orf27 |
chromosome 15 open reading frame 27 |
| chrX_-_110655306 | 0.13 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr1_-_24469602 | 0.13 |
ENST00000270800.1 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
| chr3_-_108248169 | 0.13 |
ENST00000273353.3 |
MYH15 |
myosin, heavy chain 15 |
| chr1_+_180601139 | 0.13 |
ENST00000367590.4 ENST00000367589.3 |
XPR1 |
xenotropic and polytropic retrovirus receptor 1 |
| chr2_-_227050079 | 0.13 |
ENST00000423838.1 |
AC068138.1 |
AC068138.1 |
| chr4_-_74486347 | 0.13 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr11_-_118023490 | 0.12 |
ENST00000324727.4 |
SCN4B |
sodium channel, voltage-gated, type IV, beta subunit |
| chr6_-_161695042 | 0.12 |
ENST00000366908.5 ENST00000366911.5 ENST00000366905.3 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr17_-_38956205 | 0.12 |
ENST00000306658.7 |
KRT28 |
keratin 28 |
| chr4_-_74486109 | 0.12 |
ENST00000395777.2 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr2_-_99871570 | 0.12 |
ENST00000333017.2 ENST00000409679.1 ENST00000423306.1 |
LYG2 |
lysozyme G-like 2 |
| chr12_-_89746173 | 0.12 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chrX_+_139791917 | 0.12 |
ENST00000607004.1 ENST00000370535.3 |
LINC00632 |
long intergenic non-protein coding RNA 632 |
| chr4_-_76944621 | 0.12 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr12_+_20963647 | 0.12 |
ENST00000381545.3 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr12_+_20963632 | 0.12 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr1_-_92952433 | 0.12 |
ENST00000294702.5 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr4_-_39979576 | 0.11 |
ENST00000303538.8 ENST00000503396.1 |
PDS5A |
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr11_-_129062093 | 0.11 |
ENST00000310343.9 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr12_-_86650045 | 0.11 |
ENST00000604798.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chrX_+_78003204 | 0.11 |
ENST00000435339.3 ENST00000514744.1 |
LPAR4 |
lysophosphatidic acid receptor 4 |
| chr3_-_27763803 | 0.11 |
ENST00000449599.1 |
EOMES |
eomesodermin |
| chrM_+_10758 | 0.11 |
ENST00000361381.2 |
MT-ND4 |
mitochondrially encoded NADH dehydrogenase 4 |
| chr8_+_50824233 | 0.11 |
ENST00000522124.1 |
SNTG1 |
syntrophin, gamma 1 |
| chr4_+_169013666 | 0.11 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr4_-_143227088 | 0.10 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr1_+_107683644 | 0.10 |
ENST00000370067.1 |
NTNG1 |
netrin G1 |
| chr12_+_28410128 | 0.10 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr4_-_123542224 | 0.09 |
ENST00000264497.3 |
IL21 |
interleukin 21 |
| chr1_-_150738261 | 0.09 |
ENST00000448301.2 ENST00000368985.3 |
CTSS |
cathepsin S |
| chr6_+_12290586 | 0.09 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr4_-_143226979 | 0.09 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr20_+_52105495 | 0.09 |
ENST00000439873.2 |
AL354993.1 |
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr17_-_39280419 | 0.09 |
ENST00000394014.1 |
KRTAP4-12 |
keratin associated protein 4-12 |
| chr1_+_107682629 | 0.09 |
ENST00000370074.4 ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1 |
netrin G1 |
| chr7_-_71868354 | 0.09 |
ENST00000412588.1 |
CALN1 |
calneuron 1 |
| chr10_+_32873190 | 0.09 |
ENST00000375025.4 |
C10orf68 |
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr2_+_27440229 | 0.09 |
ENST00000264705.4 ENST00000403525.1 |
CAD |
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr3_-_52090461 | 0.09 |
ENST00000296483.6 ENST00000495880.1 |
DUSP7 |
dual specificity phosphatase 7 |
| chr3_+_173116225 | 0.08 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr4_+_41614909 | 0.08 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr17_-_38938786 | 0.08 |
ENST00000301656.3 |
KRT27 |
keratin 27 |
| chrX_-_71458802 | 0.08 |
ENST00000373657.1 ENST00000334463.3 |
ERCC6L |
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr12_-_112123524 | 0.08 |
ENST00000327551.6 |
BRAP |
BRCA1 associated protein |
| chr3_-_79816965 | 0.08 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr2_-_207024134 | 0.08 |
ENST00000457011.1 ENST00000440274.1 ENST00000432169.1 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr19_+_1440838 | 0.08 |
ENST00000594262.1 |
AC027307.3 |
Uncharacterized protein |
| chr2_-_207023918 | 0.08 |
ENST00000455934.2 ENST00000449699.1 ENST00000454195.1 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr14_-_38064198 | 0.08 |
ENST00000250448.2 |
FOXA1 |
forkhead box A1 |
| chr6_+_151042224 | 0.07 |
ENST00000358517.2 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr1_-_152386732 | 0.07 |
ENST00000271835.3 |
CRNN |
cornulin |
| chr14_-_72458326 | 0.07 |
ENST00000542853.1 |
AC005477.1 |
AC005477.1 |
| chr2_-_163008903 | 0.07 |
ENST00000418842.2 ENST00000375497.3 |
GCG |
glucagon |
| chr7_-_83278322 | 0.07 |
ENST00000307792.3 |
SEMA3E |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr7_+_50348268 | 0.07 |
ENST00000438033.1 ENST00000439701.1 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
| chr20_-_29978383 | 0.07 |
ENST00000339144.3 ENST00000376321.3 |
DEFB119 |
defensin, beta 119 |
| chr6_+_12007897 | 0.07 |
ENST00000437559.1 |
RP11-456H18.2 |
RP11-456H18.2 |
| chr12_-_10978957 | 0.07 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr5_+_61874562 | 0.07 |
ENST00000334994.5 ENST00000409534.1 |
LRRC70 IPO11 |
leucine rich repeat containing 70 importin 11 |
| chr9_-_131486367 | 0.07 |
ENST00000372663.4 ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12 |
zinc finger, DHHC-type containing 12 |
| chr8_+_26150628 | 0.07 |
ENST00000523925.1 ENST00000315985.7 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr14_-_104181771 | 0.07 |
ENST00000554913.1 ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3 |
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr4_-_87028478 | 0.07 |
ENST00000515400.1 ENST00000395157.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr8_+_107460147 | 0.07 |
ENST00000442977.2 |
OXR1 |
oxidation resistance 1 |
| chr18_+_21572737 | 0.06 |
ENST00000304621.6 |
TTC39C |
tetratricopeptide repeat domain 39C |
| chr8_+_99129513 | 0.06 |
ENST00000522319.1 ENST00000401707.2 |
POP1 |
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr11_+_63606373 | 0.06 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr1_+_160160346 | 0.06 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr14_-_36988882 | 0.06 |
ENST00000498187.2 |
NKX2-1 |
NK2 homeobox 1 |
| chr18_-_33709268 | 0.06 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr12_-_52967600 | 0.06 |
ENST00000549343.1 ENST00000305620.2 |
KRT74 |
keratin 74 |
| chr6_-_9933500 | 0.06 |
ENST00000492169.1 |
OFCC1 |
orofacial cleft 1 candidate 1 |
| chr1_+_155023757 | 0.06 |
ENST00000356955.2 ENST00000449910.2 ENST00000359280.4 ENST00000360674.4 ENST00000368412.3 ENST00000355956.2 ENST00000368410.2 ENST00000271836.6 ENST00000368413.1 ENST00000531455.1 ENST00000447332.3 |
ADAM15 |
ADAM metallopeptidase domain 15 |
| chr1_+_160160283 | 0.06 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr11_+_34663913 | 0.06 |
ENST00000532302.1 |
EHF |
ets homologous factor |
| chr19_-_36304201 | 0.06 |
ENST00000301175.3 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr4_-_89951028 | 0.06 |
ENST00000506913.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr17_+_41363854 | 0.06 |
ENST00000588693.1 ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A |
transmembrane protein 106A |
| chr19_+_4402659 | 0.06 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chrX_+_107288239 | 0.06 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr16_+_30669720 | 0.06 |
ENST00000356166.6 |
FBRS |
fibrosin |
| chr14_+_22739823 | 0.06 |
ENST00000390464.2 |
TRAV38-1 |
T cell receptor alpha variable 38-1 |
| chr17_-_46716647 | 0.06 |
ENST00000608940.1 |
RP11-357H14.17 |
RP11-357H14.17 |
| chr6_-_167797887 | 0.06 |
ENST00000476779.2 ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10 |
t-complex 10 |
| chr17_+_72427477 | 0.05 |
ENST00000342648.5 ENST00000481232.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr19_+_48949030 | 0.05 |
ENST00000253237.5 |
GRWD1 |
glutamate-rich WD repeat containing 1 |
| chr13_-_44735393 | 0.05 |
ENST00000400419.1 |
SMIM2 |
small integral membrane protein 2 |
| chr20_+_30697298 | 0.05 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr21_-_32253874 | 0.05 |
ENST00000332378.4 |
KRTAP11-1 |
keratin associated protein 11-1 |
| chr11_+_34664014 | 0.05 |
ENST00000527935.1 |
EHF |
ets homologous factor |
| chr12_+_122688090 | 0.05 |
ENST00000324189.4 ENST00000546192.1 |
B3GNT4 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr7_-_130080977 | 0.05 |
ENST00000223208.5 |
CEP41 |
centrosomal protein 41kDa |
| chr4_+_88571429 | 0.05 |
ENST00000339673.6 ENST00000282479.7 |
DMP1 |
dentin matrix acidic phosphoprotein 1 |
| chr2_+_103035102 | 0.05 |
ENST00000264260.2 |
IL18RAP |
interleukin 18 receptor accessory protein |
| chr2_-_208031943 | 0.05 |
ENST00000421199.1 ENST00000457962.1 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr10_-_50970322 | 0.05 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr12_-_3862245 | 0.05 |
ENST00000252322.1 ENST00000440314.2 |
EFCAB4B |
EF-hand calcium binding domain 4B |
| chr11_+_20620946 | 0.05 |
ENST00000525748.1 |
SLC6A5 |
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr13_-_36050819 | 0.05 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr3_+_16306837 | 0.05 |
ENST00000606098.1 |
OXNAD1 |
oxidoreductase NAD-binding domain containing 1 |
| chr3_-_74570291 | 0.05 |
ENST00000263665.6 |
CNTN3 |
contactin 3 (plasmacytoma associated) |
| chr12_-_57328187 | 0.04 |
ENST00000293502.1 |
SDR9C7 |
short chain dehydrogenase/reductase family 9C, member 7 |
| chr6_+_135502501 | 0.04 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr17_+_42785976 | 0.04 |
ENST00000393547.2 ENST00000398338.3 |
DBF4B |
DBF4 homolog B (S. cerevisiae) |
| chr3_+_121774202 | 0.04 |
ENST00000469710.1 ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86 |
CD86 molecule |
| chr7_-_111032971 | 0.04 |
ENST00000450877.1 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr7_-_87342564 | 0.04 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr6_-_39693111 | 0.04 |
ENST00000373215.3 ENST00000538893.1 ENST00000287152.7 ENST00000373216.3 |
KIF6 |
kinesin family member 6 |
| chr7_+_129984630 | 0.04 |
ENST00000355388.3 ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5 |
carboxypeptidase A5 |
| chr21_-_31538971 | 0.04 |
ENST00000286808.3 |
CLDN17 |
claudin 17 |
| chr19_+_10397648 | 0.04 |
ENST00000340992.4 ENST00000393717.2 |
ICAM4 |
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
| chrX_+_134654540 | 0.04 |
ENST00000370752.4 |
DDX26B |
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr5_+_126984710 | 0.04 |
ENST00000379445.3 |
CTXN3 |
cortexin 3 |
| chr7_-_44580861 | 0.04 |
ENST00000546276.1 ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1 |
NPC1-like 1 |
| chr9_+_130026756 | 0.04 |
ENST00000314904.5 ENST00000373387.4 |
GARNL3 |
GTPase activating Rap/RanGAP domain-like 3 |
| chr3_-_12587055 | 0.04 |
ENST00000564146.3 |
C3orf83 |
chromosome 3 open reading frame 83 |
| chr17_-_38911580 | 0.04 |
ENST00000312150.4 |
KRT25 |
keratin 25 |
| chr18_+_32173276 | 0.04 |
ENST00000591816.1 ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA |
dystrobrevin, alpha |
| chrM_+_9207 | 0.04 |
ENST00000362079.2 |
MT-CO3 |
mitochondrially encoded cytochrome c oxidase III |
| chr1_-_186430222 | 0.04 |
ENST00000391997.2 |
PDC |
phosducin |
| chr10_+_115511213 | 0.03 |
ENST00000361048.1 |
PLEKHS1 |
pleckstrin homology domain containing, family S member 1 |
| chr17_-_38821373 | 0.03 |
ENST00000394052.3 |
KRT222 |
keratin 222 |
| chr12_-_14849470 | 0.03 |
ENST00000261170.3 |
GUCY2C |
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr10_-_24770632 | 0.03 |
ENST00000596413.1 |
AL353583.1 |
AL353583.1 |
| chr14_+_32798547 | 0.03 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chrX_-_18690210 | 0.03 |
ENST00000379984.3 |
RS1 |
retinoschisin 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.2 | 0.3 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.1 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 2.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 1.0 | GO:1902224 | ketone body metabolic process(GO:1902224) |
| 0.1 | 0.4 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.4 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.4 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.1 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.1 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.2 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.0 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.0 | 0.0 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.0 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.0 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.0 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.2 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.3 | GO:0071481 | cellular response to X-ray(GO:0071481) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 1.0 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 2.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |