Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
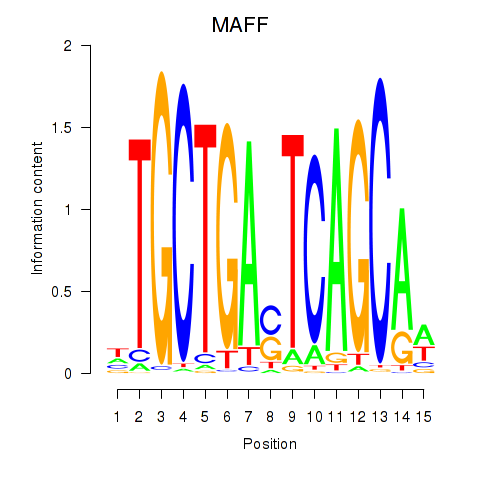
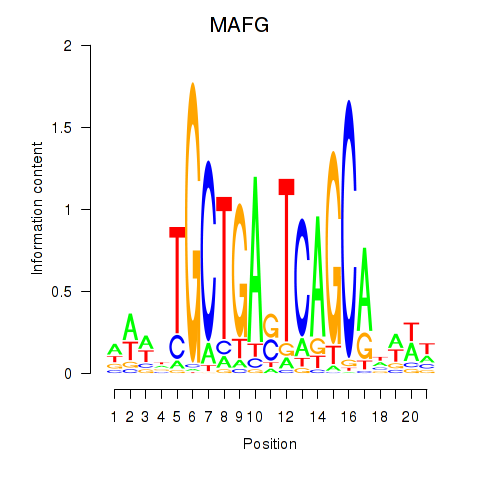
Results for MAFF_MAFG
Z-value: 1.34
Transcription factors associated with MAFF_MAFG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFF
|
ENSG00000185022.7 | MAFF |
|
MAFG
|
ENSG00000197063.6 | MAFG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAFF | hg19_v2_chr22_+_38597889_38598021 | -0.87 | 4.5e-03 | Click! |
| MAFG | hg19_v2_chr17_-_79881408_79881423 | -0.78 | 2.1e-02 | Click! |
Activity profile of MAFF_MAFG motif
Sorted Z-values of MAFF_MAFG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFF_MAFG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_100199800 | 1.48 |
ENST00000223061.5 |
PCOLCE |
procollagen C-endopeptidase enhancer |
| chr12_-_91573132 | 1.25 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr12_+_56075330 | 1.22 |
ENST00000394252.3 |
METTL7B |
methyltransferase like 7B |
| chr22_-_44708731 | 1.09 |
ENST00000381176.4 |
KIAA1644 |
KIAA1644 |
| chr3_+_45071622 | 1.02 |
ENST00000428034.1 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr11_+_75428857 | 1.00 |
ENST00000198801.5 |
MOGAT2 |
monoacylglycerol O-acyltransferase 2 |
| chr5_+_110409012 | 0.90 |
ENST00000379706.4 |
TSLP |
thymic stromal lymphopoietin |
| chr5_-_111092873 | 0.80 |
ENST00000509025.1 ENST00000515855.1 |
NREP |
neuronal regeneration related protein |
| chr5_-_111093167 | 0.77 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr15_+_33010175 | 0.76 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chrX_-_107019181 | 0.74 |
ENST00000315660.4 ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr16_+_66442411 | 0.70 |
ENST00000499966.1 |
LINC00920 |
long intergenic non-protein coding RNA 920 |
| chr12_-_47219733 | 0.67 |
ENST00000547477.1 ENST00000447411.1 ENST00000266579.4 |
SLC38A4 |
solute carrier family 38, member 4 |
| chr4_+_26585538 | 0.65 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr1_+_183774240 | 0.64 |
ENST00000360851.3 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr5_-_149535421 | 0.64 |
ENST00000261799.4 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
| chr5_-_111093081 | 0.63 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr1_+_110210644 | 0.62 |
ENST00000369831.2 ENST00000442650.1 ENST00000369827.3 ENST00000460717.3 ENST00000241337.4 ENST00000467579.3 ENST00000414179.2 ENST00000369829.2 |
GSTM2 |
glutathione S-transferase mu 2 (muscle) |
| chr8_-_91657740 | 0.57 |
ENST00000422900.1 |
TMEM64 |
transmembrane protein 64 |
| chr5_-_111093340 | 0.56 |
ENST00000508870.1 |
NREP |
neuronal regeneration related protein |
| chr17_-_76921459 | 0.55 |
ENST00000262768.7 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr20_-_45061695 | 0.54 |
ENST00000445496.2 |
ELMO2 |
engulfment and cell motility 2 |
| chr5_-_111093406 | 0.53 |
ENST00000379671.3 |
NREP |
neuronal regeneration related protein |
| chr1_-_232598163 | 0.52 |
ENST00000308942.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr2_-_190044480 | 0.49 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr4_+_74718906 | 0.47 |
ENST00000226524.3 |
PF4V1 |
platelet factor 4 variant 1 |
| chr22_+_22730353 | 0.46 |
ENST00000390296.2 |
IGLV5-45 |
immunoglobulin lambda variable 5-45 |
| chr16_-_87970122 | 0.45 |
ENST00000309893.2 |
CA5A |
carbonic anhydrase VA, mitochondrial |
| chr7_-_14026063 | 0.43 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr20_+_12989596 | 0.42 |
ENST00000434210.1 ENST00000399002.2 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
| chr1_+_110254850 | 0.41 |
ENST00000369812.5 ENST00000256593.3 ENST00000369813.1 |
GSTM5 |
glutathione S-transferase mu 5 |
| chrX_-_63005405 | 0.41 |
ENST00000374878.1 ENST00000437457.2 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr2_+_74781828 | 0.41 |
ENST00000340004.6 |
DOK1 |
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
| chr17_-_63822563 | 0.41 |
ENST00000317442.8 |
CEP112 |
centrosomal protein 112kDa |
| chr17_-_28618867 | 0.39 |
ENST00000394819.3 ENST00000577623.1 |
BLMH |
bleomycin hydrolase |
| chr17_-_28618948 | 0.39 |
ENST00000261714.6 |
BLMH |
bleomycin hydrolase |
| chrX_+_22050546 | 0.38 |
ENST00000379374.4 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr5_+_140529630 | 0.38 |
ENST00000543635.1 |
PCDHB6 |
protocadherin beta 6 |
| chr16_-_3350614 | 0.37 |
ENST00000268674.2 |
TIGD7 |
tigger transposable element derived 7 |
| chr3_-_178969403 | 0.37 |
ENST00000314235.5 ENST00000392685.2 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chrX_-_118284542 | 0.34 |
ENST00000402510.2 |
KIAA1210 |
KIAA1210 |
| chr7_-_14026123 | 0.34 |
ENST00000420159.2 ENST00000399357.3 ENST00000403527.1 |
ETV1 |
ets variant 1 |
| chr6_-_152489484 | 0.34 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr19_+_5914213 | 0.34 |
ENST00000222125.5 ENST00000452990.2 ENST00000588865.1 |
CAPS |
calcyphosine |
| chr1_-_12891264 | 0.32 |
ENST00000535591.1 ENST00000437584.1 |
PRAMEF11 |
PRAME family member 11 |
| chr5_+_139027877 | 0.32 |
ENST00000302517.3 |
CXXC5 |
CXXC finger protein 5 |
| chr12_+_32655048 | 0.31 |
ENST00000427716.2 ENST00000266482.3 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr1_-_110283138 | 0.31 |
ENST00000256594.3 |
GSTM3 |
glutathione S-transferase mu 3 (brain) |
| chr21_-_45078019 | 0.30 |
ENST00000542962.1 |
HSF2BP |
heat shock transcription factor 2 binding protein |
| chr10_+_134150835 | 0.30 |
ENST00000432555.2 |
LRRC27 |
leucine rich repeat containing 27 |
| chr9_-_95166841 | 0.30 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr22_+_44319648 | 0.30 |
ENST00000423180.2 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
| chr4_+_26585686 | 0.29 |
ENST00000505206.1 ENST00000511789.1 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr3_-_99594948 | 0.29 |
ENST00000471562.1 ENST00000495625.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr10_-_90751038 | 0.28 |
ENST00000458159.1 ENST00000415557.1 ENST00000458208.1 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr9_-_95166884 | 0.28 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr1_-_27998689 | 0.27 |
ENST00000339145.4 ENST00000362020.4 ENST00000361157.6 |
IFI6 |
interferon, alpha-inducible protein 6 |
| chr7_-_122526799 | 0.27 |
ENST00000334010.7 ENST00000313070.7 |
CADPS2 |
Ca++-dependent secretion activator 2 |
| chr5_+_140602904 | 0.27 |
ENST00000515856.2 ENST00000239449.4 |
PCDHB14 |
protocadherin beta 14 |
| chr1_+_110230412 | 0.26 |
ENST00000309851.5 ENST00000369823.2 |
GSTM1 |
glutathione S-transferase mu 1 |
| chr4_-_8073705 | 0.26 |
ENST00000514025.1 |
ABLIM2 |
actin binding LIM protein family, member 2 |
| chr6_+_80816342 | 0.26 |
ENST00000369760.4 ENST00000356489.5 ENST00000320393.6 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr3_-_141944398 | 0.25 |
ENST00000544571.1 ENST00000392993.2 |
GK5 |
glycerol kinase 5 (putative) |
| chr4_+_74347400 | 0.25 |
ENST00000226355.3 |
AFM |
afamin |
| chr1_+_89829610 | 0.25 |
ENST00000370456.4 ENST00000535065.1 |
GBP6 |
guanylate binding protein family, member 6 |
| chr11_+_62379194 | 0.25 |
ENST00000525801.1 ENST00000534093.1 |
ROM1 |
retinal outer segment membrane protein 1 |
| chr16_-_24942273 | 0.24 |
ENST00000571406.1 |
ARHGAP17 |
Rho GTPase activating protein 17 |
| chr3_-_58563094 | 0.24 |
ENST00000464064.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr19_-_53606604 | 0.24 |
ENST00000599056.1 ENST00000599247.1 ENST00000355147.5 ENST00000429604.1 ENST00000418871.1 ENST00000599637.1 |
ZNF160 |
zinc finger protein 160 |
| chr21_+_17909594 | 0.24 |
ENST00000441820.1 ENST00000602280.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr1_-_24438664 | 0.23 |
ENST00000374434.3 ENST00000330966.7 ENST00000329601.7 |
MYOM3 |
myomesin 3 |
| chr9_-_113800341 | 0.23 |
ENST00000358883.4 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr4_-_1670632 | 0.23 |
ENST00000461064.1 |
FAM53A |
family with sequence similarity 53, member A |
| chr10_-_45496336 | 0.23 |
ENST00000298298.1 |
C10orf25 |
chromosome 10 open reading frame 25 |
| chr6_-_43478239 | 0.23 |
ENST00000372441.1 |
LRRC73 |
leucine rich repeat containing 73 |
| chr3_-_99595037 | 0.23 |
ENST00000383694.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr2_-_167232484 | 0.23 |
ENST00000375387.4 ENST00000303354.6 ENST00000409672.1 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
| chrX_+_102883887 | 0.23 |
ENST00000372625.3 ENST00000372624.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr9_+_114423615 | 0.23 |
ENST00000374293.4 |
GNG10 |
guanine nucleotide binding protein (G protein), gamma 10 |
| chr17_+_66509019 | 0.23 |
ENST00000585981.1 ENST00000589480.1 ENST00000585815.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr1_+_145549203 | 0.22 |
ENST00000355594.4 ENST00000544626.1 |
ANKRD35 |
ankyrin repeat domain 35 |
| chr12_-_102455846 | 0.22 |
ENST00000545679.1 |
CCDC53 |
coiled-coil domain containing 53 |
| chr1_+_22979676 | 0.22 |
ENST00000432749.2 ENST00000314933.6 |
C1QB |
complement component 1, q subcomponent, B chain |
| chr5_+_176449684 | 0.22 |
ENST00000506693.1 ENST00000358149.3 ENST00000512315.1 ENST00000503425.1 |
ZNF346 |
zinc finger protein 346 |
| chr8_+_26240414 | 0.22 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr22_-_20461786 | 0.22 |
ENST00000426804.1 |
RIMBP3 |
RIMS binding protein 3 |
| chr9_-_113800317 | 0.22 |
ENST00000374431.3 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr17_+_260097 | 0.21 |
ENST00000360127.6 ENST00000571106.1 ENST00000491373.1 |
C17orf97 |
chromosome 17 open reading frame 97 |
| chr12_+_51318513 | 0.21 |
ENST00000332160.4 |
METTL7A |
methyltransferase like 7A |
| chr1_+_196788887 | 0.21 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr1_-_109655355 | 0.21 |
ENST00000369945.3 |
C1orf194 |
chromosome 1 open reading frame 194 |
| chr1_-_13002348 | 0.20 |
ENST00000355096.2 |
PRAMEF6 |
PRAME family member 6 |
| chr7_+_139025875 | 0.20 |
ENST00000297534.6 |
C7orf55 |
chromosome 7 open reading frame 55 |
| chr6_+_10556215 | 0.20 |
ENST00000316170.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr9_-_119449483 | 0.20 |
ENST00000288520.5 ENST00000358637.4 ENST00000341734.4 |
ASTN2 |
astrotactin 2 |
| chr22_+_22707260 | 0.20 |
ENST00000390293.1 |
IGLV5-48 |
immunoglobulin lambda variable 5-48 (non-functional) |
| chr12_+_103981044 | 0.20 |
ENST00000388887.2 |
STAB2 |
stabilin 2 |
| chr6_+_25963020 | 0.20 |
ENST00000357085.3 |
TRIM38 |
tripartite motif containing 38 |
| chr10_+_124320156 | 0.20 |
ENST00000338354.3 ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr14_-_69619823 | 0.20 |
ENST00000341516.5 |
DCAF5 |
DDB1 and CUL4 associated factor 5 |
| chr11_+_67171358 | 0.19 |
ENST00000526387.1 |
TBC1D10C |
TBC1 domain family, member 10C |
| chr17_-_76899275 | 0.19 |
ENST00000322630.2 ENST00000586713.1 |
DDC8 |
Protein DDC8 homolog |
| chr19_+_44645700 | 0.19 |
ENST00000592437.1 |
ZNF234 |
zinc finger protein 234 |
| chr10_-_44144292 | 0.19 |
ENST00000374433.2 |
ZNF32 |
zinc finger protein 32 |
| chr1_+_22351977 | 0.19 |
ENST00000420503.1 ENST00000416769.1 ENST00000404210.2 |
LINC00339 |
long intergenic non-protein coding RNA 339 |
| chr17_+_7452189 | 0.19 |
ENST00000293825.6 |
TNFSF12 |
tumor necrosis factor (ligand) superfamily, member 12 |
| chr5_+_172571445 | 0.19 |
ENST00000231668.9 ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1 |
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr17_+_66508537 | 0.18 |
ENST00000392711.1 ENST00000585427.1 ENST00000589228.1 ENST00000536854.2 ENST00000588702.1 ENST00000589309.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr19_-_58459039 | 0.18 |
ENST00000282308.3 ENST00000598928.1 |
ZNF256 |
zinc finger protein 256 |
| chr11_+_67171391 | 0.18 |
ENST00000312390.5 |
TBC1D10C |
TBC1 domain family, member 10C |
| chr6_-_167571817 | 0.18 |
ENST00000366834.1 |
GPR31 |
G protein-coupled receptor 31 |
| chrX_+_155110956 | 0.18 |
ENST00000286448.6 ENST00000262640.6 ENST00000460621.1 |
VAMP7 |
vesicle-associated membrane protein 7 |
| chr9_+_134378289 | 0.18 |
ENST00000423007.1 ENST00000404875.2 ENST00000441334.1 ENST00000341012.7 ENST00000372228.3 ENST00000402686.3 ENST00000419118.2 ENST00000541219.1 ENST00000354713.4 ENST00000418774.1 ENST00000415075.1 ENST00000448212.1 ENST00000430619.1 |
POMT1 |
protein-O-mannosyltransferase 1 |
| chr8_-_27850141 | 0.18 |
ENST00000524352.1 |
SCARA5 |
scavenger receptor class A, member 5 (putative) |
| chr17_+_18625336 | 0.18 |
ENST00000395671.4 ENST00000571542.1 ENST00000395672.2 ENST00000414850.2 ENST00000424146.2 |
TRIM16L |
tripartite motif containing 16-like |
| chr10_+_124030819 | 0.18 |
ENST00000260723.4 ENST00000368994.2 |
BTBD16 |
BTB (POZ) domain containing 16 |
| chr5_-_54468974 | 0.18 |
ENST00000381375.2 ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B |
cell division cycle 20B |
| chr12_-_46385811 | 0.18 |
ENST00000419565.2 |
SCAF11 |
SR-related CTD-associated factor 11 |
| chr9_+_131549610 | 0.18 |
ENST00000223865.8 |
TBC1D13 |
TBC1 domain family, member 13 |
| chr16_-_1429627 | 0.17 |
ENST00000248104.7 |
UNKL |
unkempt family zinc finger-like |
| chr17_+_66508154 | 0.17 |
ENST00000358598.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr9_-_104357277 | 0.17 |
ENST00000374806.1 |
PPP3R2 |
protein phosphatase 3, regulatory subunit B, beta |
| chr1_+_145301735 | 0.17 |
ENST00000605176.1 |
NBPF10 |
neuroblastoma breakpoint family, member 10 |
| chr17_-_64187973 | 0.17 |
ENST00000583358.1 ENST00000392769.2 |
CEP112 |
centrosomal protein 112kDa |
| chr6_+_126112001 | 0.17 |
ENST00000392477.2 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr7_-_86849883 | 0.17 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr12_+_10103893 | 0.17 |
ENST00000355690.4 |
CLEC12A |
C-type lectin domain family 12, member A |
| chr6_+_132455118 | 0.17 |
ENST00000458028.1 |
LINC01013 |
long intergenic non-protein coding RNA 1013 |
| chr11_-_114271139 | 0.16 |
ENST00000325636.4 |
C11orf71 |
chromosome 11 open reading frame 71 |
| chr6_-_88299678 | 0.16 |
ENST00000369536.5 |
RARS2 |
arginyl-tRNA synthetase 2, mitochondrial |
| chr17_-_47022140 | 0.16 |
ENST00000290330.3 |
SNF8 |
SNF8, ESCRT-II complex subunit |
| chr10_-_100174900 | 0.16 |
ENST00000370575.4 |
PYROXD2 |
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr5_-_115152651 | 0.16 |
ENST00000250535.4 |
CDO1 |
cysteine dioxygenase type 1 |
| chr22_-_19435209 | 0.16 |
ENST00000546308.1 ENST00000541063.1 ENST00000399568.1 ENST00000333059.5 |
HIRA C22orf39 |
histone cell cycle regulator chromosome 22 open reading frame 39 |
| chr12_-_102455902 | 0.16 |
ENST00000240079.6 |
CCDC53 |
coiled-coil domain containing 53 |
| chr10_+_90750493 | 0.16 |
ENST00000357339.2 ENST00000355279.2 |
FAS |
Fas cell surface death receptor |
| chr9_-_138391692 | 0.16 |
ENST00000429260.2 |
C9orf116 |
chromosome 9 open reading frame 116 |
| chr5_+_130506475 | 0.16 |
ENST00000379380.4 |
LYRM7 |
LYR motif containing 7 |
| chr9_-_14693417 | 0.16 |
ENST00000380916.4 |
ZDHHC21 |
zinc finger, DHHC-type containing 21 |
| chr4_+_667686 | 0.16 |
ENST00000505477.1 |
MYL5 |
myosin, light chain 5, regulatory |
| chr22_-_21905750 | 0.15 |
ENST00000433039.1 |
RIMBP3C |
RIMS binding protein 3C |
| chr1_+_85527987 | 0.15 |
ENST00000326813.8 ENST00000294664.6 ENST00000528899.1 |
WDR63 |
WD repeat domain 63 |
| chr2_+_242167319 | 0.15 |
ENST00000601871.1 |
AC104841.2 |
HCG1777198, isoform CRA_a; PRO2900; Uncharacterized protein |
| chr14_-_69619291 | 0.15 |
ENST00000554215.1 ENST00000556847.1 |
DCAF5 |
DDB1 and CUL4 associated factor 5 |
| chr16_+_446713 | 0.15 |
ENST00000397722.1 ENST00000454619.1 |
NME4 |
NME/NM23 nucleoside diphosphate kinase 4 |
| chr4_+_154125565 | 0.15 |
ENST00000338700.5 |
TRIM2 |
tripartite motif containing 2 |
| chr12_+_111051902 | 0.15 |
ENST00000397655.3 ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1 |
tectonic family member 1 |
| chr19_-_23578220 | 0.15 |
ENST00000595533.1 ENST00000397082.2 ENST00000599743.1 ENST00000300619.7 |
ZNF91 |
zinc finger protein 91 |
| chr12_+_56862301 | 0.15 |
ENST00000338146.5 |
SPRYD4 |
SPRY domain containing 4 |
| chr4_+_668348 | 0.15 |
ENST00000511290.1 |
MYL5 |
myosin, light chain 5, regulatory |
| chr8_+_96037205 | 0.15 |
ENST00000396124.4 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr17_+_60758814 | 0.15 |
ENST00000579432.1 ENST00000446119.2 |
MRC2 |
mannose receptor, C type 2 |
| chr11_-_64703354 | 0.15 |
ENST00000532246.1 ENST00000279168.2 |
GPHA2 |
glycoprotein hormone alpha 2 |
| chr11_-_77791156 | 0.15 |
ENST00000281031.4 |
NDUFC2 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr10_-_7513904 | 0.15 |
ENST00000420395.1 |
RP5-1031D4.2 |
RP5-1031D4.2 |
| chr10_+_88854926 | 0.15 |
ENST00000298784.1 ENST00000298786.4 |
FAM35A |
family with sequence similarity 35, member A |
| chr13_-_53024725 | 0.14 |
ENST00000378060.4 |
VPS36 |
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr12_-_110511424 | 0.14 |
ENST00000548191.1 |
C12orf76 |
chromosome 12 open reading frame 76 |
| chr5_+_71616188 | 0.14 |
ENST00000380639.5 ENST00000543322.1 ENST00000503868.1 ENST00000510676.2 ENST00000536805.1 |
PTCD2 |
pentatricopeptide repeat domain 2 |
| chrX_-_64196376 | 0.14 |
ENST00000447788.2 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr16_+_56716336 | 0.14 |
ENST00000394485.4 ENST00000562939.1 |
MT1X |
metallothionein 1X |
| chr2_+_201981527 | 0.14 |
ENST00000441224.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr7_-_14029515 | 0.14 |
ENST00000430479.1 ENST00000405218.2 ENST00000343495.5 |
ETV1 |
ets variant 1 |
| chr7_+_142498725 | 0.14 |
ENST00000466254.1 |
TRBC2 |
T cell receptor beta constant 2 |
| chr7_+_100551239 | 0.14 |
ENST00000319509.7 |
MUC3A |
mucin 3A, cell surface associated |
| chr13_+_108921977 | 0.14 |
ENST00000430559.1 ENST00000375887.4 |
TNFSF13B |
tumor necrosis factor (ligand) superfamily, member 13b |
| chrX_-_64196351 | 0.14 |
ENST00000374839.3 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr3_+_44771088 | 0.14 |
ENST00000396048.2 |
ZNF501 |
zinc finger protein 501 |
| chrX_-_134156502 | 0.14 |
ENST00000391440.1 |
FAM127C |
family with sequence similarity 127, member C |
| chr16_-_18801643 | 0.14 |
ENST00000322989.4 ENST00000563390.1 |
RPS15A |
ribosomal protein S15a |
| chr6_+_131894284 | 0.14 |
ENST00000368087.3 ENST00000356962.2 |
ARG1 |
arginase 1 |
| chr1_+_22979474 | 0.14 |
ENST00000509305.1 |
C1QB |
complement component 1, q subcomponent, B chain |
| chr2_-_219858123 | 0.14 |
ENST00000453769.1 ENST00000295728.2 ENST00000392096.2 |
CRYBA2 |
crystallin, beta A2 |
| chr1_-_150669500 | 0.13 |
ENST00000271732.3 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr19_-_50380536 | 0.13 |
ENST00000391832.3 ENST00000391834.2 ENST00000344175.5 |
AKT1S1 |
AKT1 substrate 1 (proline-rich) |
| chr5_+_176449726 | 0.13 |
ENST00000261948.4 ENST00000511834.1 ENST00000503039.1 |
ZNF346 |
zinc finger protein 346 |
| chr7_-_150777949 | 0.13 |
ENST00000482571.1 |
FASTK |
Fas-activated serine/threonine kinase |
| chr18_-_55470320 | 0.13 |
ENST00000536015.1 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr1_-_238054094 | 0.13 |
ENST00000366570.4 |
ZP4 |
zona pellucida glycoprotein 4 |
| chr2_+_46844290 | 0.13 |
ENST00000238892.3 |
CRIPT |
cysteine-rich PDZ-binding protein |
| chr22_-_36635684 | 0.13 |
ENST00000358502.5 |
APOL2 |
apolipoprotein L, 2 |
| chr15_+_90895471 | 0.13 |
ENST00000354377.3 ENST00000379090.5 |
ZNF774 |
zinc finger protein 774 |
| chr11_+_66276550 | 0.13 |
ENST00000419755.3 |
CTD-3074O7.11 |
Bardet-Biedl syndrome 1 protein |
| chrX_+_30260054 | 0.13 |
ENST00000378982.2 |
MAGEB4 |
melanoma antigen family B, 4 |
| chr22_-_30867973 | 0.13 |
ENST00000402286.1 ENST00000401751.1 ENST00000539629.1 ENST00000403066.1 ENST00000215812.4 |
SEC14L3 |
SEC14-like 3 (S. cerevisiae) |
| chr1_-_113249734 | 0.13 |
ENST00000484054.3 ENST00000369636.2 ENST00000369637.1 ENST00000285735.2 ENST00000369638.2 |
RHOC |
ras homolog family member C |
| chr19_+_1248547 | 0.12 |
ENST00000586757.1 ENST00000300952.2 |
MIDN |
midnolin |
| chr10_+_54074033 | 0.12 |
ENST00000373970.3 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
| chr6_-_32374900 | 0.12 |
ENST00000374995.3 ENST00000374993.1 ENST00000414363.1 ENST00000540315.1 ENST00000544175.1 ENST00000429232.2 ENST00000454136.3 ENST00000446536.2 |
BTNL2 |
butyrophilin-like 2 (MHC class II associated) |
| chr4_-_185655278 | 0.12 |
ENST00000281453.5 |
MLF1IP |
centromere protein U |
| chr22_+_23154239 | 0.12 |
ENST00000390315.2 |
IGLV3-10 |
immunoglobulin lambda variable 3-10 |
| chr6_+_46661575 | 0.12 |
ENST00000450697.1 |
TDRD6 |
tudor domain containing 6 |
| chrX_-_64196307 | 0.12 |
ENST00000545618.1 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chrX_-_55187588 | 0.12 |
ENST00000472571.2 ENST00000332132.4 ENST00000425133.2 ENST00000358460.4 |
FAM104B |
family with sequence similarity 104, member B |
| chr8_-_135522425 | 0.12 |
ENST00000521673.1 |
ZFAT |
zinc finger and AT hook domain containing |
| chr16_+_58783542 | 0.12 |
ENST00000500117.1 ENST00000565722.1 |
RP11-410D17.2 |
RP11-410D17.2 |
| chr5_-_175612149 | 0.12 |
ENST00000515403.1 |
RP11-844P9.2 |
RP11-844P9.2 |
| chr17_-_61819121 | 0.12 |
ENST00000245865.5 ENST00000375840.4 ENST00000582137.1 ENST00000579549.1 ENST00000582030.1 ENST00000584110.1 ENST00000580288.1 ENST00000336174.6 ENST00000579340.1 ENST00000580338.1 |
STRADA |
STE20-related kinase adaptor alpha |
| chr10_+_120116527 | 0.12 |
ENST00000445161.1 |
LINC00867 |
long intergenic non-protein coding RNA 867 |
| chr8_-_42623747 | 0.12 |
ENST00000534622.1 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 1.2 | GO:0070458 | cellular detoxification of nitrogen compound(GO:0070458) |
| 0.2 | 1.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.6 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 1.0 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.8 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.5 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.6 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.6 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 1.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.7 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.2 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.4 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.2 | GO:0033214 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.1 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.2 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.1 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.2 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.9 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.1 | GO:1903248 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.2 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.3 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.4 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.3 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.1 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.2 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1904344 | positive regulation of growth rate(GO:0040010) negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0019520 | pentose-phosphate shunt, oxidative branch(GO:0009051) aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.0 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0048733 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) sebaceous gland development(GO:0048733) |
| 0.0 | 0.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.4 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.1 | GO:0070269 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) pyroptosis(GO:0070269) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.0 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 1.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.2 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.0 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.2 | 0.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.4 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.3 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.7 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 1.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.5 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 2.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.0 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.8 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 2.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |