Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for MEF2C
Z-value: 0.06
Transcription factors associated with MEF2C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2C
|
ENSG00000081189.9 | MEF2C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2C | hg19_v2_chr5_-_88119580_88119605 | -0.33 | 4.2e-01 | Click! |
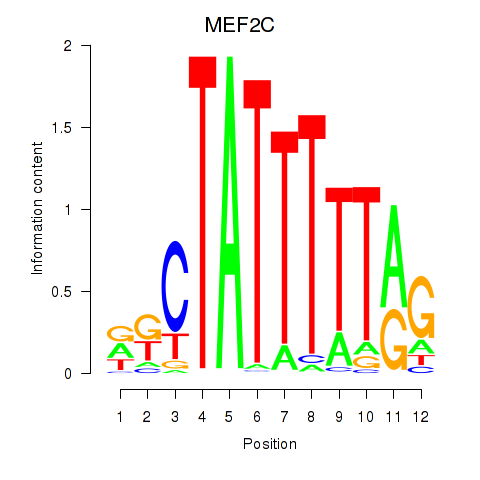
Activity profile of MEF2C motif
Sorted Z-values of MEF2C motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2C
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_186697044 | 0.46 |
ENST00000437304.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr1_-_31845914 | 0.35 |
ENST00000373713.2 |
FABP3 |
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr4_+_55095264 | 0.31 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr2_+_233497931 | 0.31 |
ENST00000264059.3 |
EFHD1 |
EF-hand domain family, member D1 |
| chr11_+_1860200 | 0.31 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr21_+_27011584 | 0.27 |
ENST00000400532.1 ENST00000480456.1 ENST00000312957.5 |
JAM2 |
junctional adhesion molecule 2 |
| chr4_-_186696425 | 0.26 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr8_+_104831472 | 0.24 |
ENST00000262231.10 ENST00000507740.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr4_-_138453606 | 0.22 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr4_+_113970772 | 0.22 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr6_+_136172820 | 0.22 |
ENST00000308191.6 |
PDE7B |
phosphodiesterase 7B |
| chr3_+_193853927 | 0.21 |
ENST00000232424.3 |
HES1 |
hes family bHLH transcription factor 1 |
| chr13_-_67802549 | 0.19 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr3_+_8543533 | 0.19 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr11_+_1860682 | 0.18 |
ENST00000381906.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr6_+_123038689 | 0.17 |
ENST00000354275.2 ENST00000368446.1 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr11_+_1860832 | 0.17 |
ENST00000252898.7 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chrX_-_142722897 | 0.17 |
ENST00000338017.4 |
SLITRK4 |
SLIT and NTRK-like family, member 4 |
| chr11_-_19223523 | 0.16 |
ENST00000265968.3 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr7_-_150946015 | 0.16 |
ENST00000262188.8 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr3_+_8543561 | 0.16 |
ENST00000397386.3 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr17_-_27949911 | 0.15 |
ENST00000492276.2 ENST00000345068.5 ENST00000584602.1 |
CORO6 |
coronin 6 |
| chr4_+_86699834 | 0.15 |
ENST00000395183.2 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr21_+_17909594 | 0.14 |
ENST00000441820.1 ENST00000602280.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr2_+_148778570 | 0.14 |
ENST00000407073.1 |
MBD5 |
methyl-CpG binding domain protein 5 |
| chr8_-_72268889 | 0.14 |
ENST00000388742.4 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr8_-_72268968 | 0.14 |
ENST00000388740.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr4_+_3465027 | 0.13 |
ENST00000389653.2 ENST00000507039.1 ENST00000340083.5 |
DOK7 |
docking protein 7 |
| chr3_+_36421826 | 0.13 |
ENST00000273183.3 |
STAC |
SH3 and cysteine rich domain |
| chr5_-_66492562 | 0.13 |
ENST00000256447.4 |
CD180 |
CD180 molecule |
| chr3_+_50316458 | 0.13 |
ENST00000316436.3 |
LSMEM2 |
leucine-rich single-pass membrane protein 2 |
| chr18_-_3219847 | 0.13 |
ENST00000261606.7 |
MYOM1 |
myomesin 1 |
| chr3_+_8543393 | 0.12 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr2_+_168043793 | 0.11 |
ENST00000409273.1 ENST00000409605.1 |
XIRP2 |
xin actin-binding repeat containing 2 |
| chr8_-_17533838 | 0.11 |
ENST00000400046.1 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chrX_+_18725758 | 0.11 |
ENST00000472826.1 ENST00000544635.1 ENST00000496075.2 |
PPEF1 |
protein phosphatase, EF-hand calcium binding domain 1 |
| chr3_+_108541545 | 0.11 |
ENST00000295756.6 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr8_+_107282368 | 0.10 |
ENST00000521369.2 |
RP11-395G23.3 |
RP11-395G23.3 |
| chr15_-_52970820 | 0.10 |
ENST00000261844.7 ENST00000399202.4 ENST00000562135.1 |
FAM214A |
family with sequence similarity 214, member A |
| chr3_+_108541608 | 0.10 |
ENST00000426646.1 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr20_-_896960 | 0.10 |
ENST00000381922.3 ENST00000546022.1 |
ANGPT4 |
angiopoietin 4 |
| chr1_-_12677714 | 0.10 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chrX_+_135279179 | 0.09 |
ENST00000370676.3 |
FHL1 |
four and a half LIM domains 1 |
| chr5_-_16509101 | 0.09 |
ENST00000399793.2 |
FAM134B |
family with sequence similarity 134, member B |
| chr4_-_25865159 | 0.09 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr4_-_57547870 | 0.09 |
ENST00000381260.3 ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX |
HOP homeobox |
| chr9_-_33402506 | 0.09 |
ENST00000377425.4 ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7 |
aquaporin 7 |
| chr3_-_52486841 | 0.09 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr4_-_57547454 | 0.09 |
ENST00000556376.2 |
HOPX |
HOP homeobox |
| chr1_+_221051699 | 0.09 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr7_+_80267973 | 0.09 |
ENST00000394788.3 ENST00000447544.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr3_+_54157480 | 0.09 |
ENST00000490478.1 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chrX_+_135278908 | 0.09 |
ENST00000539015.1 ENST00000370683.1 |
FHL1 |
four and a half LIM domains 1 |
| chr17_-_34625719 | 0.09 |
ENST00000422211.2 ENST00000542124.1 |
CCL3L1 |
chemokine (C-C motif) ligand 3-like 1 |
| chr6_-_45983549 | 0.09 |
ENST00000544153.1 |
CLIC5 |
chloride intracellular channel 5 |
| chr18_-_3220106 | 0.08 |
ENST00000356443.4 ENST00000400569.3 |
MYOM1 |
myomesin 1 |
| chr2_+_33661382 | 0.08 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr15_-_70994612 | 0.08 |
ENST00000558758.1 ENST00000379983.2 ENST00000560441.1 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr9_-_215744 | 0.08 |
ENST00000382387.2 |
C9orf66 |
chromosome 9 open reading frame 66 |
| chr3_+_154797877 | 0.08 |
ENST00000462745.1 ENST00000493237.1 |
MME |
membrane metallo-endopeptidase |
| chr3_+_127634312 | 0.08 |
ENST00000407609.3 |
KBTBD12 |
kelch repeat and BTB (POZ) domain containing 12 |
| chr11_+_1942580 | 0.08 |
ENST00000381558.1 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr2_+_220143989 | 0.08 |
ENST00000336576.5 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr9_+_116327326 | 0.08 |
ENST00000342620.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr2_+_220144052 | 0.08 |
ENST00000425450.1 ENST00000392086.4 ENST00000421532.1 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr7_+_75931861 | 0.08 |
ENST00000248553.6 |
HSPB1 |
heat shock 27kDa protein 1 |
| chr13_+_111766897 | 0.08 |
ENST00000317133.5 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr2_-_211168332 | 0.08 |
ENST00000341685.4 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr8_-_49834299 | 0.07 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr3_+_127634069 | 0.07 |
ENST00000405109.1 |
KBTBD12 |
kelch repeat and BTB (POZ) domain containing 12 |
| chr6_-_154751629 | 0.07 |
ENST00000424998.1 |
CNKSR3 |
CNKSR family member 3 |
| chr7_+_80231466 | 0.07 |
ENST00000309881.7 ENST00000534394.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr8_-_49833978 | 0.07 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr19_-_49576198 | 0.07 |
ENST00000221444.1 |
KCNA7 |
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr6_+_132129151 | 0.07 |
ENST00000360971.2 |
ENPP1 |
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr15_+_67418047 | 0.07 |
ENST00000540846.2 |
SMAD3 |
SMAD family member 3 |
| chr12_-_131200810 | 0.07 |
ENST00000536002.1 ENST00000544034.1 |
RIMBP2 RP11-662M24.2 |
RIMS binding protein 2 RP11-662M24.2 |
| chr8_+_107282389 | 0.06 |
ENST00000577661.1 ENST00000445937.1 |
RP11-395G23.3 OXR1 |
RP11-395G23.3 oxidation resistance 1 |
| chr20_-_44519839 | 0.06 |
ENST00000372518.4 |
NEURL2 |
neuralized E3 ubiquitin protein ligase 2 |
| chr17_-_34524157 | 0.06 |
ENST00000378354.4 ENST00000394484.1 |
CCL3L3 |
chemokine (C-C motif) ligand 3-like 3 |
| chr12_-_49393092 | 0.06 |
ENST00000421952.2 |
DDN |
dendrin |
| chr18_+_9708162 | 0.06 |
ENST00000578921.1 |
RAB31 |
RAB31, member RAS oncogene family |
| chr1_-_173174681 | 0.06 |
ENST00000367718.1 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr4_+_41614720 | 0.06 |
ENST00000509277.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr7_-_113559104 | 0.06 |
ENST00000284601.3 |
PPP1R3A |
protein phosphatase 1, regulatory subunit 3A |
| chr11_+_65266507 | 0.06 |
ENST00000544868.1 |
MALAT1 |
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr10_+_95517616 | 0.06 |
ENST00000371418.4 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chrX_-_15332665 | 0.06 |
ENST00000537676.1 ENST00000344384.4 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr7_+_30951461 | 0.06 |
ENST00000311813.4 |
AQP1 |
aquaporin 1 (Colton blood group) |
| chr8_-_33457453 | 0.06 |
ENST00000523956.1 ENST00000256261.4 |
DUSP26 |
dual specificity phosphatase 26 (putative) |
| chr2_+_166152283 | 0.06 |
ENST00000375427.2 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr10_+_95517566 | 0.06 |
ENST00000542308.1 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chr11_+_7597639 | 0.06 |
ENST00000533792.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr7_+_18535893 | 0.05 |
ENST00000432645.2 ENST00000441542.2 |
HDAC9 |
histone deacetylase 9 |
| chr10_+_118083919 | 0.05 |
ENST00000333254.3 |
CCDC172 |
coiled-coil domain containing 172 |
| chr4_-_122302163 | 0.05 |
ENST00000394427.2 |
QRFPR |
pyroglutamylated RFamide peptide receptor |
| chr4_-_39640513 | 0.05 |
ENST00000511809.1 ENST00000505729.1 |
SMIM14 |
small integral membrane protein 14 |
| chr21_-_42219065 | 0.05 |
ENST00000400454.1 |
DSCAM |
Down syndrome cell adhesion molecule |
| chr22_+_25595817 | 0.05 |
ENST00000215855.2 ENST00000404334.1 |
CRYBB3 |
crystallin, beta B3 |
| chr3_-_11610255 | 0.05 |
ENST00000424529.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr1_+_160051319 | 0.05 |
ENST00000368088.3 |
KCNJ9 |
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr15_+_42651691 | 0.05 |
ENST00000357568.3 ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3 |
calpain 3, (p94) |
| chr18_+_29027696 | 0.05 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr9_-_16728161 | 0.05 |
ENST00000603713.1 ENST00000603313.1 |
BNC2 |
basonuclin 2 |
| chr10_+_52751010 | 0.05 |
ENST00000373985.1 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr17_+_7184986 | 0.05 |
ENST00000317370.8 ENST00000571308.1 |
SLC2A4 |
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr20_+_43211149 | 0.05 |
ENST00000372886.1 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr17_-_45899126 | 0.05 |
ENST00000007414.3 ENST00000392507.3 |
OSBPL7 |
oxysterol binding protein-like 7 |
| chrX_-_77914825 | 0.05 |
ENST00000321110.1 |
ZCCHC5 |
zinc finger, CCHC domain containing 5 |
| chr4_-_140098339 | 0.05 |
ENST00000394235.2 |
ELF2 |
E74-like factor 2 (ets domain transcription factor) |
| chr2_+_170366203 | 0.05 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chr20_+_58251716 | 0.05 |
ENST00000355648.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr1_-_201391149 | 0.05 |
ENST00000555948.1 ENST00000556362.1 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr3_+_135741576 | 0.04 |
ENST00000334546.2 |
PPP2R3A |
protein phosphatase 2, regulatory subunit B'', alpha |
| chr2_+_145780767 | 0.04 |
ENST00000599358.1 ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr2_+_145780739 | 0.04 |
ENST00000597173.1 ENST00000602108.1 ENST00000420472.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr8_-_7309887 | 0.04 |
ENST00000458665.1 ENST00000528168.1 |
SPAG11B |
sperm associated antigen 11B |
| chr20_+_44035847 | 0.04 |
ENST00000372712.2 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr8_+_132952112 | 0.04 |
ENST00000520362.1 ENST00000519656.1 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
| chr3_+_138067314 | 0.04 |
ENST00000423968.2 |
MRAS |
muscle RAS oncogene homolog |
| chr10_+_120116527 | 0.04 |
ENST00000445161.1 |
LINC00867 |
long intergenic non-protein coding RNA 867 |
| chr3_-_192445289 | 0.04 |
ENST00000430714.1 ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12 |
fibroblast growth factor 12 |
| chr7_+_39125365 | 0.04 |
ENST00000559001.1 ENST00000464276.2 |
POU6F2 |
POU class 6 homeobox 2 |
| chr3_-_4927447 | 0.04 |
ENST00000449914.1 |
AC018816.3 |
Uncharacterized protein |
| chr6_-_76203454 | 0.04 |
ENST00000237172.7 |
FILIP1 |
filamin A interacting protein 1 |
| chr6_-_161085291 | 0.03 |
ENST00000316300.5 |
LPA |
lipoprotein, Lp(a) |
| chr3_+_35683651 | 0.03 |
ENST00000187397.4 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr7_+_129906660 | 0.03 |
ENST00000222481.4 |
CPA2 |
carboxypeptidase A2 (pancreatic) |
| chr13_+_76378305 | 0.03 |
ENST00000526371.1 ENST00000526528.1 |
LMO7 |
LIM domain 7 |
| chr3_+_138067521 | 0.03 |
ENST00000494949.1 |
MRAS |
muscle RAS oncogene homolog |
| chr13_+_97928395 | 0.03 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr10_-_45474237 | 0.03 |
ENST00000448778.1 ENST00000298295.3 |
C10orf10 |
chromosome 10 open reading frame 10 |
| chr3_-_112360116 | 0.03 |
ENST00000206423.3 ENST00000439685.2 |
CCDC80 |
coiled-coil domain containing 80 |
| chr7_-_56160625 | 0.03 |
ENST00000446428.1 ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1 |
phosphorylase kinase, gamma 1 (muscle) |
| chr3_+_159570722 | 0.03 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr3_+_138067666 | 0.03 |
ENST00000475711.1 ENST00000464896.1 |
MRAS |
muscle RAS oncogene homolog |
| chr4_+_41362796 | 0.03 |
ENST00000508501.1 ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chrX_-_109683446 | 0.03 |
ENST00000372057.1 |
AMMECR1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr7_-_56160666 | 0.03 |
ENST00000297373.2 |
PHKG1 |
phosphorylase kinase, gamma 1 (muscle) |
| chr13_+_76378357 | 0.03 |
ENST00000489941.2 ENST00000525373.1 |
LMO7 |
LIM domain 7 |
| chr5_+_126988732 | 0.03 |
ENST00000395322.3 |
CTXN3 |
cortexin 3 |
| chr2_-_151344172 | 0.03 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr10_+_124134201 | 0.03 |
ENST00000368990.3 ENST00000368988.1 ENST00000368989.2 ENST00000463663.2 |
PLEKHA1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr10_+_63661053 | 0.03 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr12_-_111358372 | 0.03 |
ENST00000548438.1 ENST00000228841.8 |
MYL2 |
myosin, light chain 2, regulatory, cardiac, slow |
| chr4_+_55095428 | 0.03 |
ENST00000508170.1 ENST00000512143.1 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr11_+_69924639 | 0.03 |
ENST00000538023.1 ENST00000398543.2 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
| chr15_+_68582544 | 0.03 |
ENST00000566008.1 |
FEM1B |
fem-1 homolog b (C. elegans) |
| chr5_-_131347501 | 0.03 |
ENST00000543479.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr2_-_179914760 | 0.03 |
ENST00000420890.2 ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141 |
coiled-coil domain containing 141 |
| chr10_-_69455873 | 0.03 |
ENST00000433211.2 |
CTNNA3 |
catenin (cadherin-associated protein), alpha 3 |
| chr16_+_12058961 | 0.03 |
ENST00000053243.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr12_-_45269430 | 0.03 |
ENST00000395487.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr14_-_61748550 | 0.03 |
ENST00000555868.1 |
TMEM30B |
transmembrane protein 30B |
| chr15_+_78830023 | 0.03 |
ENST00000599596.1 |
AC027228.1 |
HCG2005369; Uncharacterized protein |
| chr1_-_154164534 | 0.03 |
ENST00000271850.7 ENST00000368530.2 |
TPM3 |
tropomyosin 3 |
| chr3_-_39234074 | 0.03 |
ENST00000340369.3 ENST00000421646.1 ENST00000396251.1 |
XIRP1 |
xin actin-binding repeat containing 1 |
| chr9_-_74980113 | 0.03 |
ENST00000376962.5 ENST00000376960.4 ENST00000237937.3 |
ZFAND5 |
zinc finger, AN1-type domain 5 |
| chr8_-_139926236 | 0.03 |
ENST00000303045.6 ENST00000435777.1 |
COL22A1 |
collagen, type XXII, alpha 1 |
| chr4_-_88450612 | 0.03 |
ENST00000418378.1 ENST00000282470.6 |
SPARCL1 |
SPARC-like 1 (hevin) |
| chr4_-_71532339 | 0.03 |
ENST00000254801.4 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr8_+_104831554 | 0.03 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr9_+_5231413 | 0.03 |
ENST00000239316.4 |
INSL4 |
insulin-like 4 (placenta) |
| chr17_-_42200958 | 0.02 |
ENST00000336057.5 |
HDAC5 |
histone deacetylase 5 |
| chr15_+_100106126 | 0.02 |
ENST00000558812.1 ENST00000338042.6 |
MEF2A |
myocyte enhancer factor 2A |
| chr13_+_76334498 | 0.02 |
ENST00000534657.1 |
LMO7 |
LIM domain 7 |
| chr21_+_35552978 | 0.02 |
ENST00000428914.2 ENST00000609062.1 ENST00000609947.1 |
LINC00310 |
long intergenic non-protein coding RNA 310 |
| chr5_-_131347306 | 0.02 |
ENST00000296869.4 ENST00000379249.3 ENST00000379272.2 ENST00000379264.2 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr7_-_22862406 | 0.02 |
ENST00000372879.4 |
TOMM7 |
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr10_-_62704005 | 0.02 |
ENST00000337910.5 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chrX_-_15683147 | 0.02 |
ENST00000380342.3 |
TMEM27 |
transmembrane protein 27 |
| chr6_-_9939552 | 0.02 |
ENST00000460363.2 |
OFCC1 |
orofacial cleft 1 candidate 1 |
| chr17_-_42200996 | 0.02 |
ENST00000587135.1 ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5 |
histone deacetylase 5 |
| chr20_+_48807351 | 0.02 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chr7_+_143632259 | 0.02 |
ENST00000408955.2 |
OR2F2 |
olfactory receptor, family 2, subfamily F, member 2 |
| chr10_-_61495760 | 0.02 |
ENST00000395347.1 |
SLC16A9 |
solute carrier family 16, member 9 |
| chr14_+_39736582 | 0.02 |
ENST00000556148.1 ENST00000348007.3 |
CTAGE5 |
CTAGE family, member 5 |
| chr7_-_22862448 | 0.02 |
ENST00000358435.4 |
TOMM7 |
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr7_-_143956815 | 0.02 |
ENST00000493325.1 |
OR2A7 |
olfactory receptor, family 2, subfamily A, member 7 |
| chr14_+_74003818 | 0.02 |
ENST00000311148.4 |
ACOT1 |
acyl-CoA thioesterase 1 |
| chr2_+_103378472 | 0.02 |
ENST00000412401.2 |
TMEM182 |
transmembrane protein 182 |
| chr2_-_231825668 | 0.02 |
ENST00000392039.2 |
GPR55 |
G protein-coupled receptor 55 |
| chr1_-_206945830 | 0.02 |
ENST00000423557.1 |
IL10 |
interleukin 10 |
| chr7_+_18535346 | 0.02 |
ENST00000405010.3 ENST00000406451.4 ENST00000428307.2 |
HDAC9 |
histone deacetylase 9 |
| chr16_+_12059050 | 0.02 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr3_+_99357319 | 0.02 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr2_+_30569506 | 0.02 |
ENST00000421976.2 |
AC109642.1 |
AC109642.1 |
| chr1_+_145524891 | 0.02 |
ENST00000369304.3 |
ITGA10 |
integrin, alpha 10 |
| chr8_-_27115903 | 0.02 |
ENST00000350889.4 ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4 |
stathmin-like 4 |
| chr5_+_67584174 | 0.02 |
ENST00000320694.8 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr9_-_21995249 | 0.02 |
ENST00000494262.1 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr12_-_122240792 | 0.02 |
ENST00000545885.1 ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1 RHOF |
AC084018.1 ras homolog family member F (in filopodia) |
| chr8_+_1993152 | 0.02 |
ENST00000262113.4 |
MYOM2 |
myomesin 2 |
| chr14_+_78870030 | 0.02 |
ENST00000553631.1 ENST00000554719.1 |
NRXN3 |
neurexin 3 |
| chr5_-_131347583 | 0.02 |
ENST00000379255.1 ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr6_+_161123270 | 0.02 |
ENST00000366924.2 ENST00000308192.9 ENST00000418964.1 |
PLG |
plasminogen |
| chr17_-_10372875 | 0.02 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr8_+_40010989 | 0.02 |
ENST00000315792.3 |
C8orf4 |
chromosome 8 open reading frame 4 |
| chr1_-_917497 | 0.02 |
ENST00000433179.2 |
C1orf170 |
chromosome 1 open reading frame 170 |
| chr4_+_110834033 | 0.02 |
ENST00000509793.1 ENST00000265171.5 |
EGF |
epidermal growth factor |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.3 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.2 | GO:2000974 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0072019 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular response to UV-A(GO:0071492) |
| 0.0 | 0.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |