Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for MYBL1
Z-value: 2.05
Transcription factors associated with MYBL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYBL1
|
ENSG00000185697.12 | MYBL1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYBL1 | hg19_v2_chr8_-_67525473_67525518 | 0.68 | 6.3e-02 | Click! |
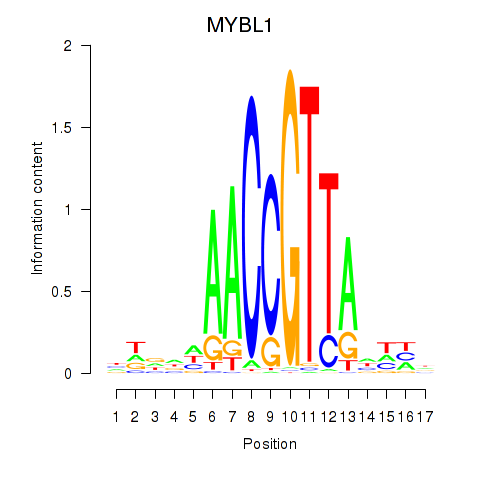
Activity profile of MYBL1 motif
Sorted Z-values of MYBL1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYBL1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_157154578 | 2.87 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr12_-_91539918 | 2.15 |
ENST00000548218.1 |
DCN |
decorin |
| chr10_+_31608054 | 1.68 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr12_-_91576561 | 1.67 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr1_+_155829286 | 1.32 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr3_+_141144963 | 1.18 |
ENST00000510726.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr12_-_47219733 | 1.14 |
ENST00000547477.1 ENST00000447411.1 ENST00000266579.4 |
SLC38A4 |
solute carrier family 38, member 4 |
| chr5_-_75919253 | 1.02 |
ENST00000296641.4 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr3_-_49314640 | 0.95 |
ENST00000436325.1 |
C3orf62 |
chromosome 3 open reading frame 62 |
| chr20_+_12989596 | 0.92 |
ENST00000434210.1 ENST00000399002.2 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
| chr5_-_146833485 | 0.85 |
ENST00000398514.3 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr2_-_190044480 | 0.81 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr4_+_123747834 | 0.78 |
ENST00000264498.3 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr8_+_104384616 | 0.77 |
ENST00000520337.1 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr11_-_27723158 | 0.76 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr6_-_56707943 | 0.75 |
ENST00000370769.4 ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST |
dystonin |
| chr19_+_44764031 | 0.74 |
ENST00000592581.1 ENST00000590668.1 ENST00000588489.1 ENST00000391958.2 |
ZNF233 |
zinc finger protein 233 |
| chr12_+_104697504 | 0.71 |
ENST00000527879.1 |
EID3 |
EP300 interacting inhibitor of differentiation 3 |
| chr13_-_33760216 | 0.69 |
ENST00000255486.4 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr10_+_91174314 | 0.69 |
ENST00000371795.4 |
IFIT5 |
interferon-induced protein with tetratricopeptide repeats 5 |
| chr3_+_141144954 | 0.65 |
ENST00000441582.2 ENST00000321464.5 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr20_+_34802295 | 0.64 |
ENST00000432603.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr12_+_56114189 | 0.62 |
ENST00000548082.1 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr12_+_56114151 | 0.61 |
ENST00000547072.1 ENST00000552930.1 ENST00000257895.5 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr1_+_210406121 | 0.57 |
ENST00000367012.3 |
SERTAD4 |
SERTA domain containing 4 |
| chr4_+_123747979 | 0.56 |
ENST00000608478.1 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr5_-_75919217 | 0.54 |
ENST00000504899.1 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr15_-_79103757 | 0.52 |
ENST00000388820.4 |
ADAMTS7 |
ADAM metallopeptidase with thrombospondin type 1 motif, 7 |
| chr12_+_53835508 | 0.51 |
ENST00000551003.1 ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13 PCBP2 |
proline rich 13 poly(rC) binding protein 2 |
| chr2_-_73460334 | 0.51 |
ENST00000258083.2 |
PRADC1 |
protease-associated domain containing 1 |
| chr19_+_37096194 | 0.50 |
ENST00000460670.1 ENST00000292928.2 ENST00000439428.1 |
ZNF382 |
zinc finger protein 382 |
| chr9_+_35658262 | 0.49 |
ENST00000378407.3 ENST00000378406.1 ENST00000426546.2 ENST00000327351.2 ENST00000421582.2 |
CCDC107 |
coiled-coil domain containing 107 |
| chr1_-_153517473 | 0.48 |
ENST00000368715.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr1_-_227505289 | 0.47 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr1_-_84464780 | 0.46 |
ENST00000260505.8 |
TTLL7 |
tubulin tyrosine ligase-like family, member 7 |
| chr13_+_32838801 | 0.46 |
ENST00000542859.1 |
FRY |
furry homolog (Drosophila) |
| chr12_+_7053172 | 0.46 |
ENST00000229281.5 |
C12orf57 |
chromosome 12 open reading frame 57 |
| chr12_+_7052974 | 0.46 |
ENST00000544681.1 ENST00000537087.1 |
C12orf57 |
chromosome 12 open reading frame 57 |
| chr1_-_145076186 | 0.44 |
ENST00000369348.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr19_-_40023450 | 0.42 |
ENST00000326282.4 |
EID2B |
EP300 interacting inhibitor of differentiation 2B |
| chr6_-_85473156 | 0.41 |
ENST00000606784.1 ENST00000606325.1 |
TBX18 |
T-box 18 |
| chr12_+_7053228 | 0.40 |
ENST00000540506.2 |
C12orf57 |
chromosome 12 open reading frame 57 |
| chr19_-_12595586 | 0.39 |
ENST00000397732.3 |
ZNF709 |
zinc finger protein 709 |
| chr20_+_5931497 | 0.39 |
ENST00000378886.2 ENST00000265187.4 |
MCM8 |
minichromosome maintenance complex component 8 |
| chr7_+_7222233 | 0.38 |
ENST00000436587.2 |
C1GALT1 |
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr10_-_28623368 | 0.38 |
ENST00000441595.2 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr19_-_52551814 | 0.37 |
ENST00000594154.1 ENST00000598745.1 ENST00000597273.1 |
ZNF432 |
zinc finger protein 432 |
| chr15_-_63449663 | 0.37 |
ENST00000439025.1 |
RPS27L |
ribosomal protein S27-like |
| chr3_-_3221358 | 0.37 |
ENST00000424814.1 ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN |
cereblon |
| chr7_+_100547156 | 0.37 |
ENST00000379458.4 |
MUC3A |
Protein LOC100131514 |
| chrX_+_70586140 | 0.37 |
ENST00000276072.3 |
TAF1 |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr1_+_97187318 | 0.36 |
ENST00000609116.1 ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2 |
polypyrimidine tract binding protein 2 |
| chr22_-_50708781 | 0.35 |
ENST00000449719.2 ENST00000330651.6 |
MAPK11 |
mitogen-activated protein kinase 11 |
| chr9_+_5510492 | 0.34 |
ENST00000397745.2 |
PDCD1LG2 |
programmed cell death 1 ligand 2 |
| chr9_+_35906176 | 0.34 |
ENST00000354323.2 |
HRCT1 |
histidine rich carboxyl terminus 1 |
| chr10_+_104614008 | 0.33 |
ENST00000369883.3 |
C10orf32 |
chromosome 10 open reading frame 32 |
| chr1_+_162467595 | 0.33 |
ENST00000538489.1 ENST00000489294.1 |
UHMK1 |
U2AF homology motif (UHM) kinase 1 |
| chr10_+_104613980 | 0.32 |
ENST00000339834.5 |
C10orf32 |
chromosome 10 open reading frame 32 |
| chr12_+_51318513 | 0.32 |
ENST00000332160.4 |
METTL7A |
methyltransferase like 7A |
| chr1_+_40974431 | 0.31 |
ENST00000296380.4 ENST00000432259.1 ENST00000418186.1 |
EXO5 |
exonuclease 5 |
| chr7_-_16685422 | 0.30 |
ENST00000306999.2 |
ANKMY2 |
ankyrin repeat and MYND domain containing 2 |
| chr10_-_33623310 | 0.29 |
ENST00000395995.1 ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1 |
neuropilin 1 |
| chr9_-_116163400 | 0.29 |
ENST00000277315.5 ENST00000448137.1 ENST00000409155.3 |
ALAD |
aminolevulinate dehydratase |
| chr4_-_6675550 | 0.29 |
ENST00000513179.1 ENST00000515205.1 |
RP11-539L10.3 |
RP11-539L10.3 |
| chr10_+_124768482 | 0.29 |
ENST00000368869.4 ENST00000358776.4 |
ACADSB |
acyl-CoA dehydrogenase, short/branched chain |
| chr8_-_91997427 | 0.29 |
ENST00000517562.2 |
RP11-122A3.2 |
chromosome 8 open reading frame 88 |
| chr11_-_47600320 | 0.28 |
ENST00000525720.1 ENST00000531067.1 ENST00000533290.1 ENST00000529499.1 ENST00000529946.1 ENST00000526005.1 ENST00000395288.2 ENST00000534239.1 |
KBTBD4 |
kelch repeat and BTB (POZ) domain containing 4 |
| chr19_-_9903666 | 0.28 |
ENST00000592587.1 |
ZNF846 |
zinc finger protein 846 |
| chr2_+_85766280 | 0.28 |
ENST00000306434.3 |
MAT2A |
methionine adenosyltransferase II, alpha |
| chr1_-_227505826 | 0.28 |
ENST00000334218.5 ENST00000366766.2 ENST00000366764.2 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr11_-_47600549 | 0.27 |
ENST00000430070.2 |
KBTBD4 |
kelch repeat and BTB (POZ) domain containing 4 |
| chr12_+_107349497 | 0.27 |
ENST00000548125.1 ENST00000280756.4 |
C12orf23 |
chromosome 12 open reading frame 23 |
| chr14_-_105420241 | 0.27 |
ENST00000557457.1 |
AHNAK2 |
AHNAK nucleoprotein 2 |
| chr10_+_124670121 | 0.26 |
ENST00000368894.1 |
FAM24A |
family with sequence similarity 24, member A |
| chr1_+_99127265 | 0.26 |
ENST00000306121.3 |
SNX7 |
sorting nexin 7 |
| chr19_+_10123925 | 0.26 |
ENST00000591589.1 ENST00000171214.1 |
RDH8 |
retinol dehydrogenase 8 (all-trans) |
| chr1_+_163291680 | 0.26 |
ENST00000450453.2 ENST00000524800.1 ENST00000442820.1 ENST00000367900.3 |
NUF2 |
NUF2, NDC80 kinetochore complex component |
| chrX_-_71792477 | 0.26 |
ENST00000421523.1 ENST00000415409.1 ENST00000373559.4 ENST00000373556.3 ENST00000373560.2 ENST00000373583.1 ENST00000429103.2 ENST00000373571.1 ENST00000373554.1 |
HDAC8 |
histone deacetylase 8 |
| chr2_-_55647057 | 0.25 |
ENST00000436346.1 |
CCDC88A |
coiled-coil domain containing 88A |
| chr2_-_55646957 | 0.25 |
ENST00000263630.8 |
CCDC88A |
coiled-coil domain containing 88A |
| chr5_-_16179884 | 0.24 |
ENST00000332432.8 |
MARCH11 |
membrane-associated ring finger (C3HC4) 11 |
| chr2_+_88047606 | 0.24 |
ENST00000359481.4 |
PLGLB2 |
plasminogen-like B2 |
| chr8_-_101157680 | 0.24 |
ENST00000428847.2 |
FBXO43 |
F-box protein 43 |
| chr1_+_211499957 | 0.24 |
ENST00000336184.2 |
TRAF5 |
TNF receptor-associated factor 5 |
| chr13_+_21714653 | 0.24 |
ENST00000382533.4 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr16_+_66442411 | 0.24 |
ENST00000499966.1 |
LINC00920 |
long intergenic non-protein coding RNA 920 |
| chr2_+_95963052 | 0.24 |
ENST00000295225.5 |
KCNIP3 |
Kv channel interacting protein 3, calsenilin |
| chr14_+_58894141 | 0.24 |
ENST00000423743.3 |
KIAA0586 |
KIAA0586 |
| chrX_+_70586082 | 0.24 |
ENST00000373790.4 ENST00000449580.1 ENST00000423759.1 |
TAF1 |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chr9_+_100069933 | 0.24 |
ENST00000529487.1 |
CCDC180 |
coiled-coil domain containing 180 |
| chr8_+_96037205 | 0.23 |
ENST00000396124.4 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr18_+_3247779 | 0.23 |
ENST00000578611.1 ENST00000583449.1 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chrX_-_153637612 | 0.23 |
ENST00000369807.1 ENST00000369808.3 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
| chr1_+_236686875 | 0.23 |
ENST00000366584.4 |
LGALS8 |
lectin, galactoside-binding, soluble, 8 |
| chr12_+_133758115 | 0.23 |
ENST00000541009.2 ENST00000592241.1 |
ZNF268 |
zinc finger protein 268 |
| chr7_-_148581360 | 0.22 |
ENST00000320356.2 ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2 |
enhancer of zeste homolog 2 (Drosophila) |
| chr9_+_104296122 | 0.22 |
ENST00000389120.3 |
RNF20 |
ring finger protein 20, E3 ubiquitin protein ligase |
| chr11_-_124981475 | 0.22 |
ENST00000532156.1 ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218 |
transmembrane protein 218 |
| chr9_+_6413317 | 0.21 |
ENST00000276893.5 ENST00000381373.3 |
UHRF2 |
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase |
| chr11_+_43380459 | 0.21 |
ENST00000299240.6 ENST00000039989.4 |
TTC17 |
tetratricopeptide repeat domain 17 |
| chr1_+_211500129 | 0.21 |
ENST00000427925.2 ENST00000261464.5 |
TRAF5 |
TNF receptor-associated factor 5 |
| chr3_+_12598563 | 0.21 |
ENST00000411987.1 ENST00000448482.1 |
MKRN2 |
makorin ring finger protein 2 |
| chr14_-_69263043 | 0.21 |
ENST00000408913.2 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
| chr12_-_76478686 | 0.21 |
ENST00000261182.8 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr9_-_21305312 | 0.20 |
ENST00000259555.4 |
IFNA5 |
interferon, alpha 5 |
| chr11_-_6677018 | 0.20 |
ENST00000299441.3 |
DCHS1 |
dachsous cadherin-related 1 |
| chr19_-_36643329 | 0.20 |
ENST00000589154.1 |
COX7A1 |
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr5_-_88179017 | 0.20 |
ENST00000514028.1 ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr20_+_1099233 | 0.20 |
ENST00000246015.4 ENST00000335877.6 ENST00000438768.2 |
PSMF1 |
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr19_-_19144243 | 0.20 |
ENST00000594445.1 ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2 |
SURP and G patch domain containing 2 |
| chr2_-_178483694 | 0.20 |
ENST00000355689.5 |
TTC30A |
tetratricopeptide repeat domain 30A |
| chr16_-_25269134 | 0.20 |
ENST00000328086.7 |
ZKSCAN2 |
zinc finger with KRAB and SCAN domains 2 |
| chr9_-_124855885 | 0.20 |
ENST00000321582.5 ENST00000373776.3 |
TTLL11 |
tubulin tyrosine ligase-like family, member 11 |
| chr2_-_86564776 | 0.20 |
ENST00000165698.5 ENST00000541910.1 ENST00000535845.1 |
REEP1 |
receptor accessory protein 1 |
| chr3_-_148804275 | 0.20 |
ENST00000392912.2 ENST00000465259.1 ENST00000310053.5 ENST00000494055.1 |
HLTF |
helicase-like transcription factor |
| chr17_-_9479128 | 0.19 |
ENST00000574431.1 |
STX8 |
syntaxin 8 |
| chr10_-_98480243 | 0.19 |
ENST00000339364.5 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
| chr12_-_66563728 | 0.19 |
ENST00000286424.7 ENST00000556010.1 ENST00000398033.4 |
TMBIM4 |
transmembrane BAX inhibitor motif containing 4 |
| chr14_+_58894103 | 0.19 |
ENST00000354386.6 ENST00000556134.1 |
KIAA0586 |
KIAA0586 |
| chr17_-_30185971 | 0.19 |
ENST00000378634.2 |
COPRS |
coordinator of PRMT5, differentiation stimulator |
| chr2_+_204103663 | 0.19 |
ENST00000356079.4 ENST00000429815.2 |
CYP20A1 |
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr12_-_89919965 | 0.19 |
ENST00000548729.1 |
POC1B-GALNT4 |
POC1B-GALNT4 readthrough |
| chr4_+_2965307 | 0.18 |
ENST00000398051.4 ENST00000503518.2 ENST00000398052.4 ENST00000345167.6 ENST00000504933.1 ENST00000442472.2 |
GRK4 |
G protein-coupled receptor kinase 4 |
| chr7_+_80275621 | 0.18 |
ENST00000426978.1 ENST00000432207.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr7_+_64254766 | 0.18 |
ENST00000307355.7 ENST00000359735.3 |
ZNF138 |
zinc finger protein 138 |
| chr1_+_99127225 | 0.18 |
ENST00000370189.5 ENST00000529992.1 |
SNX7 |
sorting nexin 7 |
| chr6_+_71377473 | 0.18 |
ENST00000370452.3 ENST00000316999.5 ENST00000370455.3 |
SMAP1 |
small ArfGAP 1 |
| chr10_-_7453445 | 0.18 |
ENST00000379713.3 ENST00000397167.1 ENST00000397160.3 |
SFMBT2 |
Scm-like with four mbt domains 2 |
| chr12_-_76478446 | 0.17 |
ENST00000393263.3 ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr17_+_46908350 | 0.17 |
ENST00000258947.3 ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2 |
calcium binding and coiled-coil domain 2 |
| chr6_+_35227449 | 0.17 |
ENST00000373953.3 ENST00000440666.2 ENST00000339411.5 |
ZNF76 |
zinc finger protein 76 |
| chr1_-_193074504 | 0.17 |
ENST00000367439.3 |
GLRX2 |
glutaredoxin 2 |
| chr10_-_23003460 | 0.17 |
ENST00000376573.4 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr1_+_45805728 | 0.17 |
ENST00000539779.1 |
TOE1 |
target of EGR1, member 1 (nuclear) |
| chr6_+_26240561 | 0.17 |
ENST00000377745.2 |
HIST1H4F |
histone cluster 1, H4f |
| chr1_-_158656488 | 0.17 |
ENST00000368147.4 |
SPTA1 |
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chrX_-_135962876 | 0.17 |
ENST00000431446.3 ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX |
RNA binding motif protein, X-linked |
| chr21_-_31852663 | 0.17 |
ENST00000390689.2 |
KRTAP19-1 |
keratin associated protein 19-1 |
| chr2_+_122513109 | 0.16 |
ENST00000389682.3 ENST00000536142.1 |
TSN |
translin |
| chr5_-_176778803 | 0.16 |
ENST00000303127.7 |
LMAN2 |
lectin, mannose-binding 2 |
| chr16_-_4784128 | 0.16 |
ENST00000592711.1 ENST00000590147.1 ENST00000304283.4 ENST00000592190.1 ENST00000589065.1 ENST00000585773.1 ENST00000450067.2 ENST00000592698.1 ENST00000586166.1 ENST00000586605.1 ENST00000592421.1 |
ANKS3 |
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr18_-_71959159 | 0.16 |
ENST00000494131.2 ENST00000397914.4 ENST00000340533.4 |
CYB5A |
cytochrome b5 type A (microsomal) |
| chr2_-_133427767 | 0.15 |
ENST00000397463.2 |
LYPD1 |
LY6/PLAUR domain containing 1 |
| chr12_+_53835383 | 0.15 |
ENST00000429243.2 |
PRR13 |
proline rich 13 |
| chr11_+_65029233 | 0.15 |
ENST00000265465.3 |
POLA2 |
polymerase (DNA directed), alpha 2, accessory subunit |
| chr1_-_19615744 | 0.15 |
ENST00000361640.4 |
AKR7A3 |
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr17_-_1420182 | 0.15 |
ENST00000421807.2 |
INPP5K |
inositol polyphosphate-5-phosphatase K |
| chr3_+_50712672 | 0.15 |
ENST00000266037.9 |
DOCK3 |
dedicator of cytokinesis 3 |
| chr7_+_99102573 | 0.15 |
ENST00000394170.2 |
ZKSCAN5 |
zinc finger with KRAB and SCAN domains 5 |
| chr9_+_139221880 | 0.15 |
ENST00000392945.3 ENST00000440944.1 |
GPSM1 |
G-protein signaling modulator 1 |
| chr2_+_208576355 | 0.15 |
ENST00000420822.1 ENST00000295414.3 ENST00000339882.5 |
CCNYL1 |
cyclin Y-like 1 |
| chr7_+_80275752 | 0.15 |
ENST00000419819.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr1_+_163291732 | 0.14 |
ENST00000271452.3 |
NUF2 |
NUF2, NDC80 kinetochore complex component |
| chr3_+_114012819 | 0.14 |
ENST00000383671.3 |
TIGIT |
T cell immunoreceptor with Ig and ITIM domains |
| chrX_+_46404928 | 0.14 |
ENST00000421685.2 ENST00000609887.1 |
ZNF674-AS1 |
ZNF674 antisense RNA 1 (head to head) |
| chr1_+_43855560 | 0.14 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr20_-_34117447 | 0.14 |
ENST00000246199.2 ENST00000424444.1 ENST00000374345.4 ENST00000444723.1 |
C20orf173 |
chromosome 20 open reading frame 173 |
| chr5_+_35617940 | 0.14 |
ENST00000282469.6 ENST00000509059.1 ENST00000356031.3 ENST00000510777.1 |
SPEF2 |
sperm flagellar 2 |
| chr16_+_524850 | 0.14 |
ENST00000450428.1 ENST00000452814.1 |
RAB11FIP3 |
RAB11 family interacting protein 3 (class II) |
| chr14_+_60558627 | 0.13 |
ENST00000317623.4 ENST00000391611.2 ENST00000406854.1 ENST00000406949.1 |
PCNXL4 |
pecanex-like 4 (Drosophila) |
| chr1_-_43855479 | 0.13 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr8_-_23315190 | 0.13 |
ENST00000356206.6 ENST00000358689.4 ENST00000417069.2 |
ENTPD4 |
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr8_+_96037255 | 0.13 |
ENST00000286687.4 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr5_-_10249990 | 0.13 |
ENST00000511437.1 ENST00000280330.8 ENST00000510047.1 |
FAM173B |
family with sequence similarity 173, member B |
| chr20_+_5986727 | 0.13 |
ENST00000378863.4 |
CRLS1 |
cardiolipin synthase 1 |
| chr1_-_151882031 | 0.13 |
ENST00000489410.1 |
THEM4 |
thioesterase superfamily member 4 |
| chr1_+_63989004 | 0.13 |
ENST00000371088.4 |
EFCAB7 |
EF-hand calcium binding domain 7 |
| chr4_-_140098339 | 0.13 |
ENST00000394235.2 |
ELF2 |
E74-like factor 2 (ets domain transcription factor) |
| chr6_+_31802685 | 0.13 |
ENST00000375639.2 ENST00000375638.3 ENST00000375635.2 ENST00000375642.2 ENST00000395789.1 |
C6orf48 |
chromosome 6 open reading frame 48 |
| chr1_+_29138654 | 0.13 |
ENST00000234961.2 |
OPRD1 |
opioid receptor, delta 1 |
| chr21_-_46221684 | 0.13 |
ENST00000330942.5 |
UBE2G2 |
ubiquitin-conjugating enzyme E2G 2 |
| chr18_-_48346415 | 0.12 |
ENST00000431965.2 ENST00000436348.2 |
MRO |
maestro |
| chr20_-_34287220 | 0.12 |
ENST00000306750.3 |
NFS1 |
NFS1 cysteine desulfurase |
| chr4_+_76649797 | 0.12 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr11_+_118938485 | 0.12 |
ENST00000300793.6 |
VPS11 |
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chr3_-_182703688 | 0.12 |
ENST00000466812.1 ENST00000487822.1 ENST00000460412.1 ENST00000469954.1 |
DCUN1D1 |
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr5_+_177435986 | 0.12 |
ENST00000398106.2 |
FAM153C |
family with sequence similarity 153, member C |
| chr12_-_27167233 | 0.12 |
ENST00000535819.1 ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
| chr18_+_77905794 | 0.12 |
ENST00000587254.1 ENST00000586421.1 |
AC139100.2 |
Uncharacterized protein |
| chr3_-_71294304 | 0.12 |
ENST00000498215.1 |
FOXP1 |
forkhead box P1 |
| chr19_+_11466167 | 0.12 |
ENST00000591608.1 |
DKFZP761J1410 |
Lipid phosphate phosphatase-related protein type 2 |
| chr7_+_156433573 | 0.12 |
ENST00000311822.8 |
RNF32 |
ring finger protein 32 |
| chr16_+_4784273 | 0.12 |
ENST00000299320.5 ENST00000586724.1 |
C16orf71 |
chromosome 16 open reading frame 71 |
| chr19_+_44556158 | 0.12 |
ENST00000434772.3 ENST00000585552.1 |
ZNF223 |
zinc finger protein 223 |
| chr12_+_53835539 | 0.12 |
ENST00000547368.1 ENST00000379786.4 ENST00000551945.1 |
PRR13 |
proline rich 13 |
| chr14_+_21423611 | 0.12 |
ENST00000304625.2 |
RNASE2 |
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) |
| chr9_+_100263912 | 0.12 |
ENST00000259365.4 |
TMOD1 |
tropomodulin 1 |
| chr7_-_2272566 | 0.11 |
ENST00000402746.1 ENST00000265854.7 ENST00000429779.1 ENST00000399654.2 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
| chr2_-_70475586 | 0.11 |
ENST00000416149.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr6_+_122793058 | 0.11 |
ENST00000392491.2 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr16_-_20753114 | 0.11 |
ENST00000396083.2 |
THUMPD1 |
THUMP domain containing 1 |
| chr22_-_36220420 | 0.11 |
ENST00000473487.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr17_-_40075219 | 0.11 |
ENST00000537919.1 ENST00000352035.2 ENST00000353196.1 ENST00000393896.2 |
ACLY |
ATP citrate lyase |
| chr22_-_39268308 | 0.11 |
ENST00000407418.3 |
CBX6 |
chromobox homolog 6 |
| chr5_+_140593509 | 0.11 |
ENST00000341948.4 |
PCDHB13 |
protocadherin beta 13 |
| chr10_-_91174215 | 0.11 |
ENST00000371837.1 |
LIPA |
lipase A, lysosomal acid, cholesterol esterase |
| chr17_+_37856299 | 0.11 |
ENST00000269571.5 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr9_+_6716478 | 0.11 |
ENST00000452643.1 |
RP11-390F4.3 |
RP11-390F4.3 |
| chr13_-_41768654 | 0.11 |
ENST00000379483.3 |
KBTBD7 |
kelch repeat and BTB (POZ) domain containing 7 |
| chr7_-_1980128 | 0.11 |
ENST00000437877.1 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.9 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.4 | 1.3 | GO:1990927 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.3 | 0.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 | 3.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 0.8 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.2 | 1.7 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 | 1.3 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.2 | 1.3 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 0.4 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.1 | 0.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.6 | GO:0036369 | transcription factor catabolic process(GO:0036369) |
| 0.1 | 0.7 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.8 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.3 | GO:0070541 | response to platinum ion(GO:0070541) cellular response to lead ion(GO:0071284) |
| 0.1 | 0.3 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.2 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.2 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.2 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.9 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.4 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.3 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.2 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.3 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.2 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.2 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.5 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 1.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.1 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.4 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0050822 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.0 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.7 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.3 | 3.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.8 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.9 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 3.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.0 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 0.8 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 1.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.9 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.6 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.3 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 0.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.6 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.2 | GO:0031071 | cysteine desulfurase activity(GO:0031071) |
| 0.1 | 0.4 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.1 | 0.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 3.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 1.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 1.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.0 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.0 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |