Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
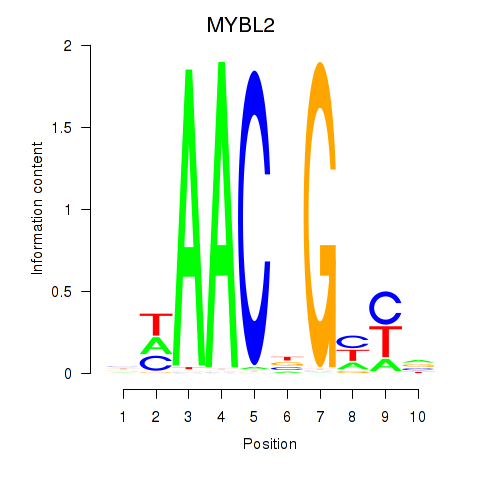
Results for MYBL2
Z-value: 0.86
Transcription factors associated with MYBL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYBL2
|
ENSG00000101057.11 | MYBL2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MYBL2 | hg19_v2_chr20_+_42295745_42295797 | 0.05 | 9.1e-01 | Click! |
Activity profile of MYBL2 motif
Sorted Z-values of MYBL2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MYBL2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_31608054 | 1.35 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr3_+_157154578 | 1.10 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr3_+_99357319 | 1.03 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr1_+_163038565 | 0.85 |
ENST00000421743.2 |
RGS4 |
regulator of G-protein signaling 4 |
| chr8_-_120685608 | 0.69 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr3_-_49314640 | 0.66 |
ENST00000436325.1 |
C3orf62 |
chromosome 3 open reading frame 62 |
| chr14_+_104394770 | 0.59 |
ENST00000409874.4 ENST00000339063.5 |
TDRD9 |
tudor domain containing 9 |
| chr2_-_68547061 | 0.57 |
ENST00000263655.3 |
CNRIP1 |
cannabinoid receptor interacting protein 1 |
| chr1_+_162602244 | 0.50 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr12_+_104697504 | 0.45 |
ENST00000527879.1 |
EID3 |
EP300 interacting inhibitor of differentiation 3 |
| chr16_+_29823427 | 0.41 |
ENST00000358758.7 ENST00000567659.1 ENST00000572820.1 |
PRRT2 |
proline-rich transmembrane protein 2 |
| chrX_-_107019181 | 0.41 |
ENST00000315660.4 ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr13_+_76210448 | 0.41 |
ENST00000377499.5 |
LMO7 |
LIM domain 7 |
| chr17_-_66951474 | 0.40 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr8_-_38326139 | 0.38 |
ENST00000335922.5 ENST00000532791.1 ENST00000397091.5 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chr9_-_79307096 | 0.38 |
ENST00000376717.2 ENST00000223609.6 ENST00000443509.2 |
PRUNE2 |
prune homolog 2 (Drosophila) |
| chr11_+_19799327 | 0.38 |
ENST00000540292.1 |
NAV2 |
neuron navigator 2 |
| chr11_+_62104897 | 0.37 |
ENST00000415229.2 ENST00000535727.1 ENST00000301776.5 |
ASRGL1 |
asparaginase like 1 |
| chr16_+_12995614 | 0.37 |
ENST00000423335.2 |
SHISA9 |
shisa family member 9 |
| chr8_-_38326119 | 0.37 |
ENST00000356207.5 ENST00000326324.6 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chr3_-_112356944 | 0.37 |
ENST00000461431.1 |
CCDC80 |
coiled-coil domain containing 80 |
| chr4_-_39640700 | 0.36 |
ENST00000295958.5 |
SMIM14 |
small integral membrane protein 14 |
| chr16_+_29823552 | 0.35 |
ENST00000300797.6 |
PRRT2 |
proline-rich transmembrane protein 2 |
| chr12_-_58329819 | 0.34 |
ENST00000551421.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr1_-_168698433 | 0.34 |
ENST00000367817.3 |
DPT |
dermatopontin |
| chr9_-_73029540 | 0.34 |
ENST00000377126.2 |
KLF9 |
Kruppel-like factor 9 |
| chr14_+_76044934 | 0.33 |
ENST00000238667.4 |
FLVCR2 |
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chr1_+_246887349 | 0.32 |
ENST00000366510.3 |
SCCPDH |
saccharopine dehydrogenase (putative) |
| chr5_+_32788945 | 0.32 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr19_-_14530143 | 0.31 |
ENST00000242776.4 |
DDX39A |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A |
| chr16_-_31076332 | 0.31 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chr9_-_79520989 | 0.28 |
ENST00000376713.3 ENST00000376718.3 ENST00000428286.1 |
PRUNE2 |
prune homolog 2 (Drosophila) |
| chr2_-_27712583 | 0.27 |
ENST00000260570.3 ENST00000359466.6 ENST00000416524.2 |
IFT172 |
intraflagellar transport 172 homolog (Chlamydomonas) |
| chr2_+_201390843 | 0.26 |
ENST00000357799.4 ENST00000409203.3 |
SGOL2 |
shugoshin-like 2 (S. pombe) |
| chr1_-_167059830 | 0.26 |
ENST00000367868.3 |
GPA33 |
glycoprotein A33 (transmembrane) |
| chr8_-_93107827 | 0.26 |
ENST00000520724.1 ENST00000518844.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr5_-_137667526 | 0.26 |
ENST00000503022.1 |
CDC25C |
cell division cycle 25C |
| chr1_-_114301503 | 0.26 |
ENST00000447664.2 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr9_+_2621798 | 0.25 |
ENST00000382100.3 |
VLDLR |
very low density lipoprotein receptor |
| chr16_-_31076273 | 0.24 |
ENST00000426488.2 |
ZNF668 |
zinc finger protein 668 |
| chr19_-_45982076 | 0.24 |
ENST00000423698.2 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr22_-_38902300 | 0.23 |
ENST00000403230.1 |
DDX17 |
DEAD (Asp-Glu-Ala-Asp) box helicase 17 |
| chr20_-_45981138 | 0.23 |
ENST00000446994.2 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr5_-_137674000 | 0.23 |
ENST00000510119.1 ENST00000513970.1 |
CDC25C |
cell division cycle 25C |
| chr12_-_58329888 | 0.23 |
ENST00000546580.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr20_-_30311703 | 0.22 |
ENST00000450273.1 ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1 |
BCL2-like 1 |
| chr17_-_56621665 | 0.22 |
ENST00000321691.3 |
C17orf47 |
chromosome 17 open reading frame 47 |
| chr16_-_4896205 | 0.22 |
ENST00000589389.1 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr12_+_79439405 | 0.21 |
ENST00000552744.1 |
SYT1 |
synaptotagmin I |
| chr12_-_76879852 | 0.21 |
ENST00000548341.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr1_-_243326612 | 0.21 |
ENST00000492145.1 ENST00000490813.1 ENST00000464936.1 |
CEP170 |
centrosomal protein 170kDa |
| chr5_-_114880533 | 0.21 |
ENST00000274457.3 |
FEM1C |
fem-1 homolog c (C. elegans) |
| chr16_+_20817761 | 0.20 |
ENST00000568046.1 ENST00000261377.6 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr17_-_67264947 | 0.20 |
ENST00000586811.1 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr17_+_7482785 | 0.20 |
ENST00000250092.6 ENST00000380498.6 ENST00000584502.1 |
CD68 |
CD68 molecule |
| chr3_-_42003479 | 0.20 |
ENST00000420927.1 |
ULK4 |
unc-51 like kinase 4 |
| chr20_+_17680587 | 0.19 |
ENST00000427254.1 ENST00000377805.3 |
BANF2 |
barrier to autointegration factor 2 |
| chr21_+_27011584 | 0.19 |
ENST00000400532.1 ENST00000480456.1 ENST00000312957.5 |
JAM2 |
junctional adhesion molecule 2 |
| chr19_-_37958318 | 0.19 |
ENST00000316950.6 ENST00000591710.1 |
ZNF569 |
zinc finger protein 569 |
| chr13_-_43566301 | 0.19 |
ENST00000398762.3 ENST00000313640.7 ENST00000313624.7 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
| chr6_+_72922590 | 0.19 |
ENST00000523963.1 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr17_-_1619535 | 0.19 |
ENST00000573075.1 ENST00000574306.1 |
MIR22HG |
MIR22 host gene (non-protein coding) |
| chr6_+_72922505 | 0.19 |
ENST00000401910.3 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr15_-_98417780 | 0.19 |
ENST00000503874.3 |
LINC00923 |
long intergenic non-protein coding RNA 923 |
| chr16_+_20817839 | 0.19 |
ENST00000348433.6 ENST00000568501.1 ENST00000566276.1 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr2_-_211036051 | 0.19 |
ENST00000418791.1 ENST00000452086.1 ENST00000281772.9 |
KANSL1L |
KAT8 regulatory NSL complex subunit 1-like |
| chrX_-_153775426 | 0.18 |
ENST00000393562.2 |
G6PD |
glucose-6-phosphate dehydrogenase |
| chr7_+_138943265 | 0.18 |
ENST00000483726.1 |
UBN2 |
ubinuclein 2 |
| chr16_-_4897266 | 0.18 |
ENST00000591451.1 ENST00000436648.5 ENST00000381983.3 ENST00000588297.1 ENST00000321919.9 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr11_-_62314268 | 0.18 |
ENST00000257247.7 ENST00000531324.1 ENST00000378024.4 |
AHNAK |
AHNAK nucleoprotein |
| chr8_-_93107696 | 0.18 |
ENST00000436581.2 ENST00000520583.1 ENST00000519061.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr10_+_94352956 | 0.17 |
ENST00000260731.3 |
KIF11 |
kinesin family member 11 |
| chr5_-_137667459 | 0.17 |
ENST00000415130.2 ENST00000356505.3 ENST00000357274.3 ENST00000348983.3 ENST00000323760.6 |
CDC25C |
cell division cycle 25C |
| chr12_+_110906169 | 0.17 |
ENST00000377673.5 |
FAM216A |
family with sequence similarity 216, member A |
| chr13_+_113656047 | 0.17 |
ENST00000375597.4 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr1_-_114301960 | 0.17 |
ENST00000369598.1 ENST00000369600.1 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr6_+_72926145 | 0.17 |
ENST00000425662.2 ENST00000453976.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr21_+_17909594 | 0.17 |
ENST00000441820.1 ENST00000602280.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr16_+_67063262 | 0.17 |
ENST00000565389.1 |
CBFB |
core-binding factor, beta subunit |
| chr11_-_9025541 | 0.17 |
ENST00000525100.1 ENST00000309166.3 ENST00000531090.1 |
NRIP3 |
nuclear receptor interacting protein 3 |
| chr2_+_30670077 | 0.17 |
ENST00000466477.1 ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1 |
lysocardiolipin acyltransferase 1 |
| chr9_+_134378289 | 0.16 |
ENST00000423007.1 ENST00000404875.2 ENST00000441334.1 ENST00000341012.7 ENST00000372228.3 ENST00000402686.3 ENST00000419118.2 ENST00000541219.1 ENST00000354713.4 ENST00000418774.1 ENST00000415075.1 ENST00000448212.1 ENST00000430619.1 |
POMT1 |
protein-O-mannosyltransferase 1 |
| chr9_+_37667978 | 0.16 |
ENST00000539465.1 |
FRMPD1 |
FERM and PDZ domain containing 1 |
| chr5_-_127674883 | 0.16 |
ENST00000507835.1 |
FBN2 |
fibrillin 2 |
| chr2_-_38830030 | 0.16 |
ENST00000410076.1 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
| chr16_+_20818020 | 0.16 |
ENST00000564274.1 ENST00000563068.1 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr17_-_5321549 | 0.16 |
ENST00000572809.1 |
NUP88 |
nucleoporin 88kDa |
| chr8_-_116680833 | 0.16 |
ENST00000220888.5 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr17_+_66511540 | 0.16 |
ENST00000588188.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr8_-_101157680 | 0.16 |
ENST00000428847.2 |
FBXO43 |
F-box protein 43 |
| chr5_+_112074029 | 0.16 |
ENST00000512211.2 |
APC |
adenomatous polyposis coli |
| chr16_+_28835437 | 0.16 |
ENST00000568266.1 |
ATXN2L |
ataxin 2-like |
| chr1_+_93646238 | 0.16 |
ENST00000448243.1 ENST00000370276.1 |
CCDC18 |
coiled-coil domain containing 18 |
| chr3_+_49027308 | 0.16 |
ENST00000383729.4 ENST00000343546.4 |
P4HTM |
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr1_-_26232522 | 0.16 |
ENST00000399728.1 |
STMN1 |
stathmin 1 |
| chr2_+_85766280 | 0.16 |
ENST00000306434.3 |
MAT2A |
methionine adenosyltransferase II, alpha |
| chr2_-_55647057 | 0.15 |
ENST00000436346.1 |
CCDC88A |
coiled-coil domain containing 88A |
| chr1_-_114301755 | 0.15 |
ENST00000393357.2 ENST00000369596.2 ENST00000446739.1 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr13_-_23949671 | 0.15 |
ENST00000402364.1 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr5_+_140810132 | 0.15 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr2_-_55646957 | 0.15 |
ENST00000263630.8 |
CCDC88A |
coiled-coil domain containing 88A |
| chr13_-_50510622 | 0.15 |
ENST00000378195.2 |
SPRYD7 |
SPRY domain containing 7 |
| chr17_-_38574169 | 0.15 |
ENST00000423485.1 |
TOP2A |
topoisomerase (DNA) II alpha 170kDa |
| chr1_-_109618566 | 0.15 |
ENST00000338366.5 |
TAF13 |
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr6_+_10521574 | 0.14 |
ENST00000495262.1 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr1_-_163172625 | 0.14 |
ENST00000527988.1 ENST00000531476.1 ENST00000530507.1 |
RGS5 |
regulator of G-protein signaling 5 |
| chr17_-_33448468 | 0.14 |
ENST00000591723.1 ENST00000593039.1 ENST00000587405.1 |
RAD51L3-RFFL RAD51D |
Uncharacterized protein RAD51 paralog D |
| chr11_+_69455855 | 0.14 |
ENST00000227507.2 ENST00000536559.1 |
CCND1 |
cyclin D1 |
| chr16_+_66586461 | 0.14 |
ENST00000264001.4 ENST00000351137.4 ENST00000345436.4 ENST00000362093.4 ENST00000417030.2 ENST00000527729.1 ENST00000532838.1 |
CKLF CKLF-CMTM1 |
chemokine-like factor CKLF-CMTM1 readthrough |
| chr11_-_67888881 | 0.14 |
ENST00000356135.5 |
CHKA |
choline kinase alpha |
| chr1_-_114302086 | 0.14 |
ENST00000369604.1 ENST00000357783.2 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr3_-_50383096 | 0.14 |
ENST00000442887.1 ENST00000360165.3 |
ZMYND10 |
zinc finger, MYND-type containing 10 |
| chr19_+_5914213 | 0.14 |
ENST00000222125.5 ENST00000452990.2 ENST00000588865.1 |
CAPS |
calcyphosine |
| chr21_+_44394620 | 0.14 |
ENST00000291547.5 |
PKNOX1 |
PBX/knotted 1 homeobox 1 |
| chrX_-_71351678 | 0.14 |
ENST00000609883.1 ENST00000545866.1 |
RGAG4 |
retrotransposon gag domain containing 4 |
| chr2_+_62132800 | 0.14 |
ENST00000538736.1 |
COMMD1 |
copper metabolism (Murr1) domain containing 1 |
| chr10_-_46168156 | 0.14 |
ENST00000374371.2 ENST00000335258.7 |
ZFAND4 |
zinc finger, AN1-type domain 4 |
| chr3_+_42642106 | 0.14 |
ENST00000232978.8 |
NKTR |
natural killer-tumor recognition sequence |
| chr5_-_180288248 | 0.14 |
ENST00000512132.1 ENST00000506439.1 ENST00000502412.1 ENST00000359141.6 |
ZFP62 |
ZFP62 zinc finger protein |
| chr13_-_60737898 | 0.14 |
ENST00000377908.2 ENST00000400319.1 ENST00000400320.1 ENST00000267215.4 |
DIAPH3 |
diaphanous-related formin 3 |
| chr3_-_138048682 | 0.14 |
ENST00000383180.2 |
NME9 |
NME/NM23 family member 9 |
| chr1_-_85156216 | 0.14 |
ENST00000342203.3 ENST00000370612.4 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr17_+_14204389 | 0.14 |
ENST00000360954.2 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
| chr12_-_46384334 | 0.14 |
ENST00000369367.3 ENST00000266589.6 ENST00000395453.2 ENST00000395454.2 |
SCAF11 |
SR-related CTD-associated factor 11 |
| chr13_-_107220455 | 0.13 |
ENST00000400198.3 |
ARGLU1 |
arginine and glutamate rich 1 |
| chr20_+_43160458 | 0.13 |
ENST00000372889.1 ENST00000372887.1 ENST00000372882.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr1_+_197881592 | 0.13 |
ENST00000367391.1 ENST00000367390.3 |
LHX9 |
LIM homeobox 9 |
| chr17_-_47287928 | 0.13 |
ENST00000507680.1 |
GNGT2 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr19_-_1812230 | 0.13 |
ENST00000587160.2 ENST00000539485.1 ENST00000310127.6 ENST00000526092.2 |
ATP8B3 |
ATPase, aminophospholipid transporter, class I, type 8B, member 3 |
| chr15_-_91537723 | 0.13 |
ENST00000394249.3 ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1 |
protein regulator of cytokinesis 1 |
| chr19_+_49867181 | 0.13 |
ENST00000597546.1 |
DKKL1 |
dickkopf-like 1 |
| chr4_+_2627159 | 0.13 |
ENST00000382839.3 ENST00000324666.5 ENST00000545951.1 ENST00000502458.1 ENST00000505311.1 |
FAM193A |
family with sequence similarity 193, member A |
| chr2_-_69870747 | 0.13 |
ENST00000409068.1 |
AAK1 |
AP2 associated kinase 1 |
| chr1_-_27998689 | 0.13 |
ENST00000339145.4 ENST00000362020.4 ENST00000361157.6 |
IFI6 |
interferon, alpha-inducible protein 6 |
| chr6_+_43543942 | 0.13 |
ENST00000372226.1 ENST00000443535.1 |
POLH |
polymerase (DNA directed), eta |
| chr4_-_7873981 | 0.13 |
ENST00000360265.4 |
AFAP1 |
actin filament associated protein 1 |
| chr19_+_852291 | 0.13 |
ENST00000263621.1 |
ELANE |
elastase, neutrophil expressed |
| chr5_+_140792614 | 0.13 |
ENST00000398610.2 |
PCDHGA10 |
protocadherin gamma subfamily A, 10 |
| chr1_-_241799232 | 0.12 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chr15_+_67430339 | 0.12 |
ENST00000439724.3 |
SMAD3 |
SMAD family member 3 |
| chr14_-_74181106 | 0.12 |
ENST00000316836.3 |
PNMA1 |
paraneoplastic Ma antigen 1 |
| chr2_-_227664474 | 0.12 |
ENST00000305123.5 |
IRS1 |
insulin receptor substrate 1 |
| chr10_-_98480243 | 0.12 |
ENST00000339364.5 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
| chr17_-_79869077 | 0.12 |
ENST00000570391.1 |
PCYT2 |
phosphate cytidylyltransferase 2, ethanolamine |
| chr17_+_46908350 | 0.12 |
ENST00000258947.3 ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2 |
calcium binding and coiled-coil domain 2 |
| chr3_-_123680246 | 0.12 |
ENST00000488653.2 |
CCDC14 |
coiled-coil domain containing 14 |
| chr2_+_62132781 | 0.12 |
ENST00000311832.5 |
COMMD1 |
copper metabolism (Murr1) domain containing 1 |
| chr6_+_43543864 | 0.12 |
ENST00000372236.4 ENST00000535400.1 |
POLH |
polymerase (DNA directed), eta |
| chrX_-_19689106 | 0.12 |
ENST00000379716.1 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr12_+_4671352 | 0.12 |
ENST00000542744.1 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr1_-_68698222 | 0.12 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr2_-_152830441 | 0.12 |
ENST00000534999.1 ENST00000397327.2 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr17_+_62503147 | 0.12 |
ENST00000553412.1 |
CEP95 |
centrosomal protein 95kDa |
| chr5_+_135496675 | 0.12 |
ENST00000507637.1 |
SMAD5 |
SMAD family member 5 |
| chr14_+_54863667 | 0.11 |
ENST00000335183.6 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
| chr19_+_36266433 | 0.11 |
ENST00000314737.5 |
ARHGAP33 |
Rho GTPase activating protein 33 |
| chr22_+_45705728 | 0.11 |
ENST00000441876.2 |
FAM118A |
family with sequence similarity 118, member A |
| chrX_-_15333736 | 0.11 |
ENST00000380470.3 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr10_+_35415719 | 0.11 |
ENST00000474362.1 ENST00000374721.3 |
CREM |
cAMP responsive element modulator |
| chr10_+_18240834 | 0.11 |
ENST00000377371.3 ENST00000539911.1 |
SLC39A12 |
solute carrier family 39 (zinc transporter), member 12 |
| chr1_-_68698197 | 0.11 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr17_-_79869004 | 0.11 |
ENST00000573927.1 ENST00000331285.3 ENST00000572157.1 |
PCYT2 |
phosphate cytidylyltransferase 2, ethanolamine |
| chr11_+_65769946 | 0.11 |
ENST00000533166.1 |
BANF1 |
barrier to autointegration factor 1 |
| chr2_+_175260451 | 0.11 |
ENST00000458563.1 ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3 |
secernin 3 |
| chr17_+_30677136 | 0.11 |
ENST00000394670.4 ENST00000321233.6 ENST00000394673.2 ENST00000341711.6 ENST00000579634.1 ENST00000580759.1 ENST00000342555.6 ENST00000577908.1 ENST00000394679.5 ENST00000582165.1 |
ZNF207 |
zinc finger protein 207 |
| chr20_-_5485166 | 0.11 |
ENST00000589201.1 ENST00000379053.4 |
LINC00654 |
long intergenic non-protein coding RNA 654 |
| chr2_+_175260514 | 0.11 |
ENST00000424069.1 ENST00000427038.1 |
SCRN3 |
secernin 3 |
| chr16_+_19535235 | 0.11 |
ENST00000565376.2 ENST00000396208.2 |
CCP110 |
centriolar coiled coil protein 110kDa |
| chr4_+_128982430 | 0.11 |
ENST00000512292.1 ENST00000508819.1 |
LARP1B |
La ribonucleoprotein domain family, member 1B |
| chr1_+_174843548 | 0.11 |
ENST00000478442.1 ENST00000465412.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr1_+_61548225 | 0.11 |
ENST00000371187.3 |
NFIA |
nuclear factor I/A |
| chrM_-_14670 | 0.11 |
ENST00000361681.2 |
MT-ND6 |
mitochondrially encoded NADH dehydrogenase 6 |
| chrX_+_129473859 | 0.11 |
ENST00000424447.1 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr1_+_211499957 | 0.11 |
ENST00000336184.2 |
TRAF5 |
TNF receptor-associated factor 5 |
| chr10_-_27389392 | 0.11 |
ENST00000376087.4 |
ANKRD26 |
ankyrin repeat domain 26 |
| chr17_-_40075219 | 0.11 |
ENST00000537919.1 ENST00000352035.2 ENST00000353196.1 ENST00000393896.2 |
ACLY |
ATP citrate lyase |
| chr16_-_70285797 | 0.11 |
ENST00000435634.1 |
EXOSC6 |
exosome component 6 |
| chr19_-_53606604 | 0.11 |
ENST00000599056.1 ENST00000599247.1 ENST00000355147.5 ENST00000429604.1 ENST00000418871.1 ENST00000599637.1 |
ZNF160 |
zinc finger protein 160 |
| chr14_+_54863682 | 0.11 |
ENST00000543789.2 ENST00000442975.2 ENST00000458126.2 ENST00000556102.2 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
| chr16_+_67062996 | 0.11 |
ENST00000561924.2 |
CBFB |
core-binding factor, beta subunit |
| chr17_-_38978847 | 0.11 |
ENST00000269576.5 |
KRT10 |
keratin 10 |
| chr22_-_23922410 | 0.11 |
ENST00000249053.3 |
IGLL1 |
immunoglobulin lambda-like polypeptide 1 |
| chr17_-_4167142 | 0.11 |
ENST00000570535.1 ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
| chr22_-_21905120 | 0.11 |
ENST00000331505.5 |
RIMBP3C |
RIMS binding protein 3C |
| chr13_+_35516390 | 0.11 |
ENST00000540320.1 ENST00000400445.3 ENST00000310336.4 |
NBEA |
neurobeachin |
| chr1_+_150480576 | 0.10 |
ENST00000346569.6 |
ECM1 |
extracellular matrix protein 1 |
| chr9_+_34652164 | 0.10 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chr16_+_30662360 | 0.10 |
ENST00000542965.2 |
PRR14 |
proline rich 14 |
| chr8_-_62602327 | 0.10 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr19_+_10765699 | 0.10 |
ENST00000590009.1 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
| chr10_-_101945771 | 0.10 |
ENST00000370408.2 ENST00000407654.3 |
ERLIN1 |
ER lipid raft associated 1 |
| chr5_-_159846399 | 0.10 |
ENST00000297151.4 |
SLU7 |
SLU7 splicing factor homolog (S. cerevisiae) |
| chr12_+_107168342 | 0.10 |
ENST00000392837.4 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
| chr15_+_89010923 | 0.10 |
ENST00000353598.6 |
MRPS11 |
mitochondrial ribosomal protein S11 |
| chr17_+_76210367 | 0.10 |
ENST00000592734.1 ENST00000587746.1 |
BIRC5 |
baculoviral IAP repeat containing 5 |
| chr5_-_16179884 | 0.10 |
ENST00000332432.8 |
MARCH11 |
membrane-associated ring finger (C3HC4) 11 |
| chr1_-_243418621 | 0.10 |
ENST00000366544.1 ENST00000366543.1 |
CEP170 |
centrosomal protein 170kDa |
| chr16_+_30662050 | 0.10 |
ENST00000568754.1 |
PRR14 |
proline rich 14 |
| chr4_+_148538517 | 0.10 |
ENST00000296582.3 ENST00000508208.1 |
TMEM184C |
transmembrane protein 184C |
| chr14_+_54863739 | 0.10 |
ENST00000541304.1 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.2 | 1.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 1.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.4 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.3 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.3 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.4 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 0.1 | GO:0061508 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.2 | GO:0039526 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host apoptotic process(GO:0039526) |
| 0.0 | 0.7 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.1 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.2 | GO:1903238 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.2 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.6 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0060008 | Sertoli cell differentiation(GO:0060008) Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0033242 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.3 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.4 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.0 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:0090260 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 | 0.8 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.2 | GO:0090083 | inclusion body assembly(GO:0070841) regulation of inclusion body assembly(GO:0090083) negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.5 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.0 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 1.0 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0060136 | enucleate erythrocyte differentiation(GO:0043353) embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.0 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.0 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.0 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.3 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.2 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) high-density lipoprotein particle remodeling(GO:0034375) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.3 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 0.5 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.2 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 1.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.4 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 1.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.4 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.3 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.3 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.2 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.0 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 1.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.1 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 1.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.0 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |