Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
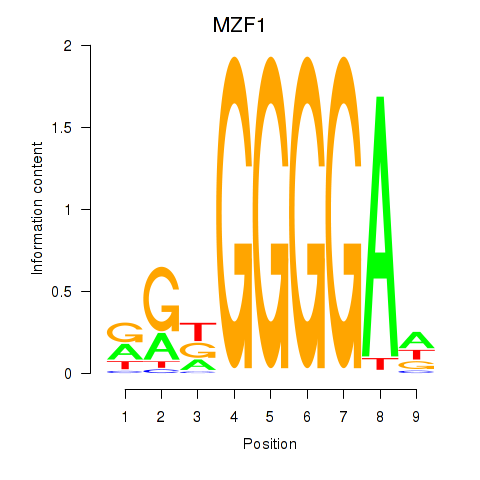
Results for MZF1
Z-value: 1.46
Transcription factors associated with MZF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MZF1
|
ENSG00000099326.4 | MZF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MZF1 | hg19_v2_chr19_-_59084922_59084947 | 0.69 | 5.6e-02 | Click! |
Activity profile of MZF1 motif
Sorted Z-values of MZF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MZF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_31608054 | 3.26 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr1_+_159141397 | 3.00 |
ENST00000368124.4 ENST00000368125.4 ENST00000416746.1 |
CADM3 |
cell adhesion molecule 3 |
| chr12_-_91576561 | 2.42 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr12_-_91576429 | 2.20 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr15_-_48937982 | 2.17 |
ENST00000316623.5 |
FBN1 |
fibrillin 1 |
| chr12_-_91576750 | 2.09 |
ENST00000228329.5 ENST00000303320.3 ENST00000052754.5 |
DCN |
decorin |
| chr5_+_82767284 | 1.76 |
ENST00000265077.3 |
VCAN |
versican |
| chr6_-_46293378 | 1.56 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr3_+_110790590 | 1.37 |
ENST00000485303.1 |
PVRL3 |
poliovirus receptor-related 3 |
| chr4_+_145567297 | 1.33 |
ENST00000434550.2 |
HHIP |
hedgehog interacting protein |
| chr5_-_81046904 | 1.32 |
ENST00000515395.1 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr11_+_113930291 | 1.22 |
ENST00000335953.4 |
ZBTB16 |
zinc finger and BTB domain containing 16 |
| chr5_-_81046841 | 1.21 |
ENST00000509013.2 ENST00000505980.1 ENST00000509053.1 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr5_+_82767583 | 1.20 |
ENST00000512590.2 ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN |
versican |
| chr11_-_35547151 | 1.18 |
ENST00000378878.3 ENST00000529303.1 ENST00000278360.3 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr11_+_86511549 | 1.12 |
ENST00000533902.2 |
PRSS23 |
protease, serine, 23 |
| chr5_+_82767487 | 1.12 |
ENST00000343200.5 ENST00000342785.4 |
VCAN |
versican |
| chr5_-_81046922 | 1.07 |
ENST00000514493.1 ENST00000320672.4 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr4_-_57547870 | 1.02 |
ENST00000381260.3 ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX |
HOP homeobox |
| chr3_+_157154578 | 1.02 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr2_+_217498105 | 0.99 |
ENST00000233809.4 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr13_+_88324870 | 0.98 |
ENST00000325089.6 |
SLITRK5 |
SLIT and NTRK-like family, member 5 |
| chr7_-_14029515 | 0.96 |
ENST00000430479.1 ENST00000405218.2 ENST00000343495.5 |
ETV1 |
ets variant 1 |
| chr2_+_33172012 | 0.93 |
ENST00000404816.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr17_+_4618734 | 0.93 |
ENST00000571206.1 |
ARRB2 |
arrestin, beta 2 |
| chr2_-_45236540 | 0.91 |
ENST00000303077.6 |
SIX2 |
SIX homeobox 2 |
| chrX_-_135849484 | 0.87 |
ENST00000370620.1 ENST00000535227.1 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr9_+_17134980 | 0.87 |
ENST00000380647.3 |
CNTLN |
centlein, centrosomal protein |
| chr11_-_2160180 | 0.83 |
ENST00000381406.4 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr1_+_155829286 | 0.83 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr11_-_96076334 | 0.82 |
ENST00000524717.1 |
MAML2 |
mastermind-like 2 (Drosophila) |
| chr17_+_39969183 | 0.82 |
ENST00000321562.4 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr2_-_175499294 | 0.82 |
ENST00000392547.2 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr1_+_162602244 | 0.81 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr1_-_153518270 | 0.80 |
ENST00000354332.4 ENST00000368716.4 |
S100A4 |
S100 calcium binding protein A4 |
| chr20_+_34742650 | 0.80 |
ENST00000373945.1 ENST00000338074.2 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chrX_-_19140575 | 0.79 |
ENST00000357991.3 ENST00000356606.4 |
GPR64 |
G protein-coupled receptor 64 |
| chr12_+_53443963 | 0.79 |
ENST00000546602.1 ENST00000552570.1 ENST00000549700.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr17_-_36904437 | 0.78 |
ENST00000585100.1 ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2 |
polycomb group ring finger 2 |
| chrX_-_70474499 | 0.77 |
ENST00000353904.2 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr1_+_211432700 | 0.77 |
ENST00000452621.2 |
RCOR3 |
REST corepressor 3 |
| chrX_-_19140677 | 0.74 |
ENST00000357544.3 ENST00000379869.3 ENST00000360279.4 ENST00000379873.2 ENST00000379878.3 ENST00000354791.3 ENST00000379876.1 |
GPR64 |
G protein-coupled receptor 64 |
| chr4_+_126237554 | 0.74 |
ENST00000394329.3 |
FAT4 |
FAT atypical cadherin 4 |
| chr17_-_48278983 | 0.73 |
ENST00000225964.5 |
COL1A1 |
collagen, type I, alpha 1 |
| chr17_-_53800217 | 0.72 |
ENST00000424486.2 |
TMEM100 |
transmembrane protein 100 |
| chr12_+_1738363 | 0.71 |
ENST00000397196.2 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
| chr2_+_219264466 | 0.70 |
ENST00000273062.2 |
CTDSP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr12_-_56106060 | 0.69 |
ENST00000452168.2 |
ITGA7 |
integrin, alpha 7 |
| chr9_+_4985016 | 0.68 |
ENST00000539801.1 |
JAK2 |
Janus kinase 2 |
| chr17_-_76921459 | 0.65 |
ENST00000262768.7 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr7_+_8008418 | 0.64 |
ENST00000223145.5 |
GLCCI1 |
glucocorticoid induced transcript 1 |
| chr12_+_53443680 | 0.64 |
ENST00000314250.6 ENST00000451358.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr1_+_78470530 | 0.61 |
ENST00000370763.5 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr3_+_141103634 | 0.59 |
ENST00000507722.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr3_+_8543393 | 0.59 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr15_-_82338460 | 0.58 |
ENST00000558133.1 ENST00000329713.4 |
MEX3B |
mex-3 RNA binding family member B |
| chr10_+_21823079 | 0.57 |
ENST00000377100.3 ENST00000377072.3 ENST00000446906.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr6_-_152957944 | 0.57 |
ENST00000423061.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr5_-_127873659 | 0.57 |
ENST00000262464.4 |
FBN2 |
fibrillin 2 |
| chr1_+_221051699 | 0.56 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr3_-_52002403 | 0.56 |
ENST00000490063.1 ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4 |
poly(rC) binding protein 4 |
| chr7_-_14029283 | 0.54 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
| chr10_+_21823243 | 0.54 |
ENST00000307729.7 ENST00000377091.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr4_+_129730839 | 0.54 |
ENST00000511647.1 |
PHF17 |
jade family PHD finger 1 |
| chr17_-_42277203 | 0.54 |
ENST00000587097.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chrX_+_102883887 | 0.53 |
ENST00000372625.3 ENST00000372624.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr14_-_65569057 | 0.53 |
ENST00000555419.1 ENST00000341653.2 |
MAX |
MYC associated factor X |
| chr3_+_148447887 | 0.53 |
ENST00000475347.1 ENST00000474935.1 ENST00000461609.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr5_-_124080203 | 0.53 |
ENST00000504926.1 |
ZNF608 |
zinc finger protein 608 |
| chr13_-_44361025 | 0.53 |
ENST00000261488.6 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
| chrX_-_70474377 | 0.53 |
ENST00000373978.1 ENST00000373981.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr6_-_137113604 | 0.53 |
ENST00000359015.4 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
| chr14_-_65569244 | 0.52 |
ENST00000557277.1 ENST00000556892.1 |
MAX |
MYC associated factor X |
| chr19_-_49371711 | 0.52 |
ENST00000355496.5 ENST00000263265.6 |
PLEKHA4 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr5_+_140729649 | 0.51 |
ENST00000523390.1 |
PCDHGB1 |
protocadherin gamma subfamily B, 1 |
| chr20_+_33292068 | 0.51 |
ENST00000374810.3 ENST00000374809.2 ENST00000451665.1 |
TP53INP2 |
tumor protein p53 inducible nuclear protein 2 |
| chr1_+_154975258 | 0.50 |
ENST00000417934.2 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
| chr2_-_72374948 | 0.50 |
ENST00000546307.1 ENST00000474509.1 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr17_+_55334364 | 0.50 |
ENST00000322684.3 ENST00000579590.1 |
MSI2 |
musashi RNA-binding protein 2 |
| chr4_+_129730947 | 0.50 |
ENST00000452328.2 ENST00000504089.1 |
PHF17 |
jade family PHD finger 1 |
| chr14_+_21538517 | 0.49 |
ENST00000298693.3 |
ARHGEF40 |
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr4_+_129730779 | 0.48 |
ENST00000226319.6 |
PHF17 |
jade family PHD finger 1 |
| chrX_+_64808248 | 0.48 |
ENST00000609672.1 |
MSN |
moesin |
| chr16_+_30907927 | 0.48 |
ENST00000279804.2 ENST00000395019.3 |
CTF1 |
cardiotrophin 1 |
| chrX_-_62974941 | 0.48 |
ENST00000374872.1 ENST00000253401.6 ENST00000374870.4 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr5_+_133861790 | 0.48 |
ENST00000395003.1 |
PHF15 |
jade family PHD finger 2 |
| chr11_+_47279504 | 0.47 |
ENST00000441012.2 ENST00000437276.1 ENST00000436029.1 ENST00000467728.1 ENST00000405853.3 |
NR1H3 |
nuclear receptor subfamily 1, group H, member 3 |
| chr6_-_31514333 | 0.47 |
ENST00000376151.4 |
ATP6V1G2 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr6_-_52860171 | 0.46 |
ENST00000370963.4 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr1_+_145516560 | 0.46 |
ENST00000537888.1 |
PEX11B |
peroxisomal biogenesis factor 11 beta |
| chr17_-_56065484 | 0.46 |
ENST00000581208.1 |
VEZF1 |
vascular endothelial zinc finger 1 |
| chr3_+_8543561 | 0.46 |
ENST00000397386.3 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr22_-_39190116 | 0.46 |
ENST00000406622.1 ENST00000216068.4 ENST00000406199.3 |
SUN2 DNAL4 |
Sad1 and UNC84 domain containing 2 dynein, axonemal, light chain 4 |
| chr1_+_211432775 | 0.45 |
ENST00000419091.2 |
RCOR3 |
REST corepressor 3 |
| chr1_-_212873267 | 0.45 |
ENST00000243440.1 |
BATF3 |
basic leucine zipper transcription factor, ATF-like 3 |
| chr20_-_45981138 | 0.45 |
ENST00000446994.2 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr1_+_150245177 | 0.44 |
ENST00000369098.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr14_+_21538429 | 0.44 |
ENST00000298694.4 ENST00000555038.1 |
ARHGEF40 |
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr1_+_145516252 | 0.44 |
ENST00000369306.3 |
PEX11B |
peroxisomal biogenesis factor 11 beta |
| chr21_+_17961006 | 0.43 |
ENST00000602323.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr11_-_6426635 | 0.43 |
ENST00000608645.1 ENST00000608394.1 ENST00000529519.1 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr14_+_74815116 | 0.43 |
ENST00000256362.4 |
VRTN |
vertebrae development associated |
| chr12_+_6875519 | 0.43 |
ENST00000389462.4 ENST00000540874.1 ENST00000309083.6 |
PTMS |
parathymosin |
| chr2_+_74685413 | 0.43 |
ENST00000233615.2 |
WBP1 |
WW domain binding protein 1 |
| chr7_+_143079000 | 0.43 |
ENST00000392910.2 |
ZYX |
zyxin |
| chr1_-_161102367 | 0.43 |
ENST00000464113.1 |
DEDD |
death effector domain containing |
| chr19_+_33166313 | 0.42 |
ENST00000334176.3 |
RGS9BP |
regulator of G protein signaling 9 binding protein |
| chr5_-_180288248 | 0.41 |
ENST00000512132.1 ENST00000506439.1 ENST00000502412.1 ENST00000359141.6 |
ZFP62 |
ZFP62 zinc finger protein |
| chr1_+_211433275 | 0.40 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr19_+_8478154 | 0.40 |
ENST00000381035.4 ENST00000595142.1 ENST00000601724.1 ENST00000393944.1 ENST00000215555.2 ENST00000601283.1 ENST00000595213.1 |
MARCH2 |
membrane-associated ring finger (C3HC4) 2, E3 ubiquitin protein ligase |
| chr3_-_120170052 | 0.40 |
ENST00000295633.3 |
FSTL1 |
follistatin-like 1 |
| chr5_-_115177247 | 0.40 |
ENST00000500945.2 |
ATG12 |
autophagy related 12 |
| chr1_+_32084794 | 0.40 |
ENST00000373705.1 |
HCRTR1 |
hypocretin (orexin) receptor 1 |
| chrX_+_54834791 | 0.40 |
ENST00000218439.4 ENST00000375058.1 ENST00000375060.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr11_-_46142615 | 0.40 |
ENST00000529734.1 ENST00000323180.6 |
PHF21A |
PHD finger protein 21A |
| chr10_+_76586348 | 0.39 |
ENST00000372724.1 ENST00000287239.4 ENST00000372714.1 |
KAT6B |
K(lysine) acetyltransferase 6B |
| chr17_+_36873677 | 0.39 |
ENST00000471200.1 |
MLLT6 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr17_+_79990058 | 0.39 |
ENST00000584341.1 |
RAC3 |
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
| chrX_-_70474910 | 0.39 |
ENST00000373988.1 ENST00000373998.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr19_+_35629702 | 0.39 |
ENST00000351325.4 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr14_-_23822061 | 0.39 |
ENST00000397260.3 |
SLC22A17 |
solute carrier family 22, member 17 |
| chr15_-_43785274 | 0.38 |
ENST00000413546.1 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr17_-_36956155 | 0.38 |
ENST00000269554.3 |
PIP4K2B |
phosphatidylinositol-5-phosphate 4-kinase, type II, beta |
| chrX_+_54834159 | 0.38 |
ENST00000375053.2 ENST00000347546.4 ENST00000375062.4 |
MAGED2 |
melanoma antigen family D, 2 |
| chr12_+_59989918 | 0.38 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr3_+_9944492 | 0.38 |
ENST00000383814.3 ENST00000454190.2 ENST00000454992.1 |
IL17RE |
interleukin 17 receptor E |
| chr15_-_37392703 | 0.38 |
ENST00000382766.2 ENST00000444725.1 |
MEIS2 |
Meis homeobox 2 |
| chr19_+_16435625 | 0.38 |
ENST00000248071.5 ENST00000592003.1 |
KLF2 |
Kruppel-like factor 2 |
| chr12_+_94542459 | 0.38 |
ENST00000258526.4 |
PLXNC1 |
plexin C1 |
| chrX_-_70473957 | 0.38 |
ENST00000373984.3 ENST00000314425.5 ENST00000373982.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr10_-_88854518 | 0.37 |
ENST00000277865.4 |
GLUD1 |
glutamate dehydrogenase 1 |
| chr5_+_140767452 | 0.37 |
ENST00000519479.1 |
PCDHGB4 |
protocadherin gamma subfamily B, 4 |
| chr1_+_145470504 | 0.37 |
ENST00000323397.4 |
ANKRD34A |
ankyrin repeat domain 34A |
| chr14_-_23822080 | 0.37 |
ENST00000397267.1 ENST00000354772.3 |
SLC22A17 |
solute carrier family 22, member 17 |
| chr4_+_113970772 | 0.37 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr2_+_46769798 | 0.37 |
ENST00000238738.4 |
RHOQ |
ras homolog family member Q |
| chr2_+_45878790 | 0.36 |
ENST00000306156.3 |
PRKCE |
protein kinase C, epsilon |
| chr21_-_44496441 | 0.36 |
ENST00000359624.3 ENST00000352178.5 |
CBS |
cystathionine-beta-synthase |
| chr12_+_59989791 | 0.36 |
ENST00000552432.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_-_155177677 | 0.36 |
ENST00000368378.3 ENST00000541990.1 ENST00000457183.2 |
THBS3 |
thrombospondin 3 |
| chr3_+_8543533 | 0.36 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chrX_+_54834004 | 0.36 |
ENST00000375068.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr7_+_90338712 | 0.36 |
ENST00000265741.3 ENST00000406263.1 |
CDK14 |
cyclin-dependent kinase 14 |
| chr12_-_48500085 | 0.36 |
ENST00000549518.1 |
SENP1 |
SUMO1/sentrin specific peptidase 1 |
| chr19_+_35630022 | 0.36 |
ENST00000589209.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr8_-_95907423 | 0.36 |
ENST00000396133.3 ENST00000308108.4 |
CCNE2 |
cyclin E2 |
| chr3_+_9944303 | 0.36 |
ENST00000421412.1 ENST00000295980.3 |
IL17RE |
interleukin 17 receptor E |
| chr6_+_25963020 | 0.35 |
ENST00000357085.3 |
TRIM38 |
tripartite motif containing 38 |
| chr22_+_24322322 | 0.35 |
ENST00000215780.5 ENST00000402588.3 |
GSTT2 |
glutathione S-transferase theta 2 |
| chr20_+_34680620 | 0.35 |
ENST00000430276.1 ENST00000373950.2 ENST00000452261.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr2_-_200322723 | 0.35 |
ENST00000417098.1 |
SATB2 |
SATB homeobox 2 |
| chr9_+_125273081 | 0.35 |
ENST00000335302.5 |
OR1J2 |
olfactory receptor, family 1, subfamily J, member 2 |
| chr12_+_79258444 | 0.35 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr1_+_201617264 | 0.35 |
ENST00000367296.4 |
NAV1 |
neuron navigator 1 |
| chr16_+_222846 | 0.35 |
ENST00000251595.6 ENST00000397806.1 |
HBA2 |
hemoglobin, alpha 2 |
| chr13_-_96296944 | 0.34 |
ENST00000361396.2 ENST00000376829.2 |
DZIP1 |
DAZ interacting zinc finger protein 1 |
| chr2_-_219925189 | 0.34 |
ENST00000295731.6 |
IHH |
indian hedgehog |
| chr10_+_181418 | 0.34 |
ENST00000403354.1 ENST00000381607.4 ENST00000402736.1 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
| chr10_-_98480243 | 0.34 |
ENST00000339364.5 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
| chr17_-_56621665 | 0.34 |
ENST00000321691.3 |
C17orf47 |
chromosome 17 open reading frame 47 |
| chr3_-_114343039 | 0.33 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr1_+_97188188 | 0.33 |
ENST00000541987.1 |
PTBP2 |
polypyrimidine tract binding protein 2 |
| chr8_-_121824374 | 0.33 |
ENST00000517992.1 |
SNTB1 |
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr17_-_73267281 | 0.33 |
ENST00000578305.1 ENST00000325102.8 |
MIF4GD |
MIF4G domain containing |
| chr17_-_73267214 | 0.33 |
ENST00000580717.1 ENST00000577542.1 ENST00000579612.1 ENST00000245551.5 |
MIF4GD |
MIF4G domain containing |
| chr7_-_23510086 | 0.33 |
ENST00000258729.3 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
| chr7_-_28220354 | 0.33 |
ENST00000283928.5 |
JAZF1 |
JAZF zinc finger 1 |
| chr17_-_73267304 | 0.33 |
ENST00000579297.1 ENST00000580571.1 |
MIF4GD |
MIF4G domain containing |
| chr6_+_42952237 | 0.33 |
ENST00000485511.1 ENST00000394110.3 ENST00000472118.1 ENST00000461010.1 |
PPP2R5D |
protein phosphatase 2, regulatory subunit B', delta |
| chr8_-_72756667 | 0.33 |
ENST00000325509.4 |
MSC |
musculin |
| chr11_-_46142505 | 0.33 |
ENST00000524497.1 ENST00000418153.2 |
PHF21A |
PHD finger protein 21A |
| chr1_-_167906277 | 0.32 |
ENST00000271373.4 |
MPC2 |
mitochondrial pyruvate carrier 2 |
| chr19_-_11347173 | 0.32 |
ENST00000587656.1 |
DOCK6 |
dedicator of cytokinesis 6 |
| chr12_+_79258547 | 0.32 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr19_+_926000 | 0.32 |
ENST00000263620.3 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
| chr11_+_86511569 | 0.32 |
ENST00000441050.1 |
PRSS23 |
protease, serine, 23 |
| chr17_+_4613918 | 0.32 |
ENST00000574954.1 ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2 |
arrestin, beta 2 |
| chr9_+_2015335 | 0.32 |
ENST00000349721.2 ENST00000357248.2 ENST00000450198.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_90679145 | 0.31 |
ENST00000265138.3 |
ARRDC3 |
arrestin domain containing 3 |
| chr12_+_1100449 | 0.31 |
ENST00000360905.4 |
ERC1 |
ELKS/RAB6-interacting/CAST family member 1 |
| chr11_-_7985193 | 0.31 |
ENST00000328600.2 |
NLRP10 |
NLR family, pyrin domain containing 10 |
| chr1_+_154975110 | 0.31 |
ENST00000535420.1 ENST00000368426.3 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
| chr11_+_64002292 | 0.31 |
ENST00000426086.2 |
VEGFB |
vascular endothelial growth factor B |
| chr2_-_175547571 | 0.31 |
ENST00000409415.3 ENST00000359761.3 ENST00000272746.5 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr22_-_31741757 | 0.31 |
ENST00000215919.3 |
PATZ1 |
POZ (BTB) and AT hook containing zinc finger 1 |
| chr7_-_14028488 | 0.31 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr11_-_66445219 | 0.31 |
ENST00000525754.1 ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B |
RNA binding motif protein 4B |
| chr1_-_154943212 | 0.30 |
ENST00000368445.5 ENST00000448116.2 ENST00000368449.4 |
SHC1 |
SHC (Src homology 2 domain containing) transforming protein 1 |
| chrX_-_13835147 | 0.30 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr5_+_49962495 | 0.30 |
ENST00000515175.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr17_+_55333876 | 0.30 |
ENST00000284073.2 |
MSI2 |
musashi RNA-binding protein 2 |
| chr10_-_102089729 | 0.30 |
ENST00000465680.2 |
PKD2L1 |
polycystic kidney disease 2-like 1 |
| chr5_+_137774706 | 0.30 |
ENST00000378339.2 ENST00000254901.5 ENST00000506158.1 |
REEP2 |
receptor accessory protein 2 |
| chr1_-_154943002 | 0.30 |
ENST00000606391.1 |
SHC1 |
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr1_+_6845578 | 0.29 |
ENST00000467404.2 ENST00000439411.2 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr15_-_37392086 | 0.29 |
ENST00000561208.1 |
MEIS2 |
Meis homeobox 2 |
| chr5_+_173315283 | 0.29 |
ENST00000265085.5 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
| chr16_+_21610797 | 0.29 |
ENST00000358154.3 |
METTL9 |
methyltransferase like 9 |
| chr18_+_7946941 | 0.29 |
ENST00000444013.1 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
| chr11_-_66336060 | 0.29 |
ENST00000310325.5 |
CTSF |
cathepsin F |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.4 | 3.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.4 | 1.7 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.4 | 3.8 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 1.5 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.3 | 0.8 | GO:1905154 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.3 | 0.8 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.3 | 1.3 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 0.7 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.2 | 0.7 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 4.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 0.8 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.2 | 0.6 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.2 | 0.5 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.2 | 1.0 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.2 | 1.3 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.2 | 0.5 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.2 | 0.8 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.4 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.7 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 1.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.1 | GO:0061419 | positive regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061419) |
| 0.1 | 0.7 | GO:1902724 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 0.5 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.6 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.9 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 1.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.7 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.1 | 0.3 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.3 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 1.0 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.8 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.5 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.1 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.1 | 0.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.8 | GO:2000467 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 | 0.1 | GO:0072054 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.1 | 0.2 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.5 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 | 0.4 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.4 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.8 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.1 | 0.3 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 3.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.5 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.1 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 0.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 0.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.5 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.4 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 1.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.3 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.5 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.2 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.2 | GO:0007418 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 | 0.3 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.1 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.3 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 0.1 | 0.3 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.1 | 0.4 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 0.4 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.4 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.2 | GO:0061428 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 1.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.3 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.8 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.2 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.2 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 1.0 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.0 | 0.1 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 1.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.5 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.9 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.8 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.1 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.1 | GO:2000564 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.1 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.9 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.4 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0003186 | tricuspid valve morphogenesis(GO:0003186) |
| 0.0 | 0.4 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 1.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.1 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.3 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.1 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.2 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 1.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:2001074 | regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.0 | 0.4 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.0 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.3 | GO:0071468 | cellular response to acidic pH(GO:0071468) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.0 | GO:0021544 | subpallium development(GO:0021544) |
| 0.0 | 0.5 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.4 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.9 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.4 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.4 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.2 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.4 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.2 | GO:0007204 | positive regulation of cytosolic calcium ion concentration(GO:0007204) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0071455 | cellular response to hyperoxia(GO:0071455) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.4 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.5 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.4 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:1902307 | positive regulation of sodium ion transmembrane transport(GO:1902307) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.0 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.0 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.0 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.2 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.4 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 3.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.7 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.6 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.6 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.3 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.1 | 0.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 5.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.9 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 2.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.5 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 2.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 2.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.5 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 1.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 1.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.3 | 2.9 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 0.3 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.3 | 1.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 0.9 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 0.7 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 0.5 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.2 | 0.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 0.8 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 4.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.4 | GO:0098809 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.1 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.4 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 0.6 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 1.5 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.6 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.5 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.6 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.6 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.7 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.6 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.4 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 5.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 3.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.2 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.1 | 0.2 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 0.2 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.1 | 0.2 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.1 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.2 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.2 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.2 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 1.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 1.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 1.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 3.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 2.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.0 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 0.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 3.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 3.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.5 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 3.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 2.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 1.7 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |