Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for NFATC1
Z-value: 1.21
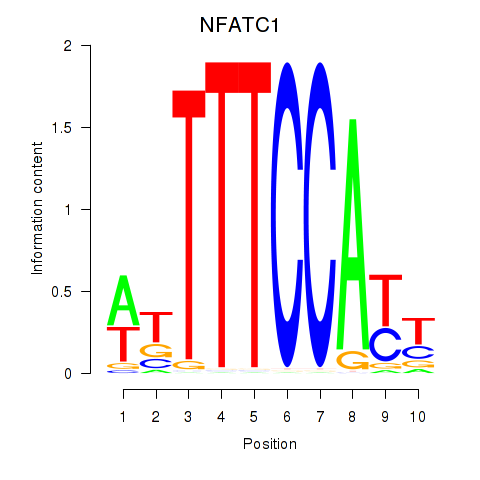
Transcription factors associated with NFATC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC1
|
ENSG00000131196.13 | NFATC1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC1 | hg19_v2_chr18_+_77155856_77155939 | -0.78 | 2.1e-02 | Click! |
Activity profile of NFATC1 motif
Sorted Z-values of NFATC1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_92413353 | 5.31 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr14_-_92413727 | 4.28 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr2_+_189839046 | 2.33 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr12_-_91573132 | 2.07 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr12_-_91573316 | 1.97 |
ENST00000393155.1 |
DCN |
decorin |
| chr15_+_33010175 | 1.24 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chrX_+_152760397 | 1.19 |
ENST00000331595.4 ENST00000431891.1 |
BGN |
biglycan |
| chr14_-_30396802 | 1.14 |
ENST00000415220.2 |
PRKD1 |
protein kinase D1 |
| chr13_-_67804445 | 1.06 |
ENST00000456367.1 ENST00000377861.3 ENST00000544246.1 |
PCDH9 |
protocadherin 9 |
| chr18_-_25616519 | 1.05 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr5_-_111093759 | 0.89 |
ENST00000509979.1 ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP |
neuronal regeneration related protein |
| chr11_+_19798964 | 0.84 |
ENST00000527559.2 |
NAV2 |
neuron navigator 2 |
| chr11_+_19799327 | 0.82 |
ENST00000540292.1 |
NAV2 |
neuron navigator 2 |
| chr16_+_15528332 | 0.78 |
ENST00000566490.1 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr2_-_145278475 | 0.66 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr14_+_21538517 | 0.66 |
ENST00000298693.3 |
ARHGEF40 |
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr9_-_20622478 | 0.65 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr1_+_162602244 | 0.65 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr14_-_30396948 | 0.63 |
ENST00000331968.5 |
PRKD1 |
protein kinase D1 |
| chr8_-_13372395 | 0.61 |
ENST00000276297.4 ENST00000511869.1 |
DLC1 |
deleted in liver cancer 1 |
| chr8_-_13372253 | 0.60 |
ENST00000316609.5 |
DLC1 |
deleted in liver cancer 1 |
| chr15_+_93443419 | 0.58 |
ENST00000557381.1 ENST00000420239.2 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr3_-_71632894 | 0.55 |
ENST00000493089.1 |
FOXP1 |
forkhead box P1 |
| chr12_-_91573249 | 0.55 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr1_-_153517473 | 0.55 |
ENST00000368715.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr5_+_32788945 | 0.54 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr5_+_140729649 | 0.54 |
ENST00000523390.1 |
PCDHGB1 |
protocadherin gamma subfamily B, 1 |
| chrX_+_9431324 | 0.52 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr3_+_69812877 | 0.51 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr17_-_29648761 | 0.51 |
ENST00000247270.3 ENST00000462804.2 |
EVI2A |
ecotropic viral integration site 2A |
| chr5_+_148521046 | 0.51 |
ENST00000326685.7 ENST00000356541.3 ENST00000309868.7 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr5_+_148521136 | 0.51 |
ENST00000506113.1 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr12_-_92536433 | 0.50 |
ENST00000551563.2 ENST00000546975.1 ENST00000549802.1 |
C12orf79 |
chromosome 12 open reading frame 79 |
| chr5_-_158526693 | 0.48 |
ENST00000380654.4 |
EBF1 |
early B-cell factor 1 |
| chr10_+_94590910 | 0.47 |
ENST00000371547.4 |
EXOC6 |
exocyst complex component 6 |
| chr8_+_77593448 | 0.46 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr8_+_77593474 | 0.45 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr5_-_41510725 | 0.45 |
ENST00000328457.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr8_-_131399110 | 0.45 |
ENST00000521426.1 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr13_+_32838801 | 0.45 |
ENST00000542859.1 |
FRY |
furry homolog (Drosophila) |
| chr5_-_41510656 | 0.44 |
ENST00000377801.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr5_-_158526756 | 0.43 |
ENST00000313708.6 ENST00000517373.1 |
EBF1 |
early B-cell factor 1 |
| chr11_-_102401469 | 0.42 |
ENST00000260227.4 |
MMP7 |
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr14_-_89883412 | 0.42 |
ENST00000557258.1 |
FOXN3 |
forkhead box N3 |
| chr3_+_148447887 | 0.42 |
ENST00000475347.1 ENST00000474935.1 ENST00000461609.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr2_+_172950227 | 0.41 |
ENST00000341900.6 |
DLX1 |
distal-less homeobox 1 |
| chr8_-_122653630 | 0.41 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr2_-_40679186 | 0.41 |
ENST00000406785.2 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chrX_-_63005405 | 0.39 |
ENST00000374878.1 ENST00000437457.2 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr10_+_105036909 | 0.39 |
ENST00000369849.4 |
INA |
internexin neuronal intermediate filament protein, alpha |
| chr18_-_77276057 | 0.39 |
ENST00000597412.1 |
AC018445.1 |
Uncharacterized protein |
| chr11_+_65190245 | 0.37 |
ENST00000499732.1 ENST00000501122.2 ENST00000601801.1 |
NEAT1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr1_+_79115503 | 0.37 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr11_-_27723158 | 0.37 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr1_+_86046433 | 0.36 |
ENST00000451137.2 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
| chr5_+_140762268 | 0.36 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr2_-_127963343 | 0.35 |
ENST00000335247.7 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr9_-_73483958 | 0.35 |
ENST00000377101.1 ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr1_+_22351977 | 0.34 |
ENST00000420503.1 ENST00000416769.1 ENST00000404210.2 |
LINC00339 |
long intergenic non-protein coding RNA 339 |
| chr8_+_70404996 | 0.34 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr5_+_139781393 | 0.31 |
ENST00000360839.2 ENST00000297183.6 ENST00000421134.1 ENST00000394723.3 ENST00000511151.1 |
ANKHD1 |
ankyrin repeat and KH domain containing 1 |
| chr1_+_61548374 | 0.31 |
ENST00000485903.2 ENST00000371185.2 ENST00000371184.2 |
NFIA |
nuclear factor I/A |
| chr12_+_26111823 | 0.31 |
ENST00000381352.3 ENST00000535907.1 ENST00000405154.2 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr10_-_92681033 | 0.30 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr12_+_6881678 | 0.30 |
ENST00000441671.2 ENST00000203629.2 |
LAG3 |
lymphocyte-activation gene 3 |
| chr8_+_38831683 | 0.30 |
ENST00000302495.4 |
HTRA4 |
HtrA serine peptidase 4 |
| chr11_-_58612168 | 0.30 |
ENST00000287275.1 |
GLYATL2 |
glycine-N-acyltransferase-like 2 |
| chr15_+_100347228 | 0.29 |
ENST00000559714.1 ENST00000560059.1 |
CTD-2054N24.2 |
Uncharacterized protein |
| chr11_+_131240373 | 0.29 |
ENST00000374791.3 ENST00000436745.1 |
NTM |
neurotrimin |
| chr3_-_127541194 | 0.29 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr3_+_151986709 | 0.28 |
ENST00000495875.2 ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr17_+_32582293 | 0.28 |
ENST00000580907.1 ENST00000225831.4 |
CCL2 |
chemokine (C-C motif) ligand 2 |
| chr2_+_11817713 | 0.28 |
ENST00000449576.2 |
LPIN1 |
lipin 1 |
| chr11_-_58611957 | 0.28 |
ENST00000532258.1 |
GLYATL2 |
glycine-N-acyltransferase-like 2 |
| chr2_+_46926326 | 0.27 |
ENST00000394861.2 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr3_+_152017924 | 0.27 |
ENST00000465907.2 ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr7_-_100493744 | 0.27 |
ENST00000428317.1 ENST00000441605.1 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr14_-_24977457 | 0.26 |
ENST00000250378.3 ENST00000206446.4 |
CMA1 |
chymase 1, mast cell |
| chr22_+_38071615 | 0.26 |
ENST00000215909.5 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
| chr12_+_32260085 | 0.26 |
ENST00000548411.1 ENST00000281474.5 ENST00000551086.1 |
BICD1 |
bicaudal D homolog 1 (Drosophila) |
| chr2_+_46926048 | 0.26 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr1_-_150780757 | 0.26 |
ENST00000271651.3 |
CTSK |
cathepsin K |
| chr5_-_73937244 | 0.25 |
ENST00000302351.4 ENST00000510316.1 ENST00000508331.1 |
ENC1 |
ectodermal-neural cortex 1 (with BTB domain) |
| chr10_-_104597286 | 0.25 |
ENST00000369887.3 |
CYP17A1 |
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr5_+_139781445 | 0.25 |
ENST00000532219.1 ENST00000394722.3 |
ANKHD1-EIF4EBP3 ANKHD1 |
ANKHD1-EIF4EBP3 readthrough ankyrin repeat and KH domain containing 1 |
| chr1_-_27816556 | 0.25 |
ENST00000536657.1 |
WASF2 |
WAS protein family, member 2 |
| chr3_-_178976996 | 0.25 |
ENST00000485523.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr7_+_20687017 | 0.25 |
ENST00000258738.6 |
ABCB5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr3_+_69134080 | 0.24 |
ENST00000273258.3 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr12_-_10605929 | 0.24 |
ENST00000347831.5 ENST00000359151.3 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr5_+_140810132 | 0.23 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr6_-_15663198 | 0.23 |
ENST00000338950.5 ENST00000511762.2 ENST00000355917.3 ENST00000344537.5 |
DTNBP1 |
dystrobrevin binding protein 1 |
| chr4_-_83765613 | 0.23 |
ENST00000503937.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr14_+_51026743 | 0.23 |
ENST00000358385.6 ENST00000357032.3 ENST00000354525.4 |
ATL1 |
atlastin GTPase 1 |
| chr6_+_132891461 | 0.23 |
ENST00000275198.1 |
TAAR6 |
trace amine associated receptor 6 |
| chr17_-_47287928 | 0.23 |
ENST00000507680.1 |
GNGT2 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr22_+_40573921 | 0.23 |
ENST00000454349.2 ENST00000335727.9 |
TNRC6B |
trinucleotide repeat containing 6B |
| chr1_-_169599353 | 0.23 |
ENST00000367793.2 ENST00000367794.2 ENST00000367792.2 ENST00000367791.2 ENST00000367788.2 |
SELP |
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr7_-_29186008 | 0.23 |
ENST00000396276.3 ENST00000265394.5 |
CPVL |
carboxypeptidase, vitellogenic-like |
| chr5_+_71014990 | 0.22 |
ENST00000296777.4 |
CARTPT |
CART prepropeptide |
| chr12_-_13529642 | 0.22 |
ENST00000318426.2 |
C12orf36 |
chromosome 12 open reading frame 36 |
| chr19_-_44388116 | 0.22 |
ENST00000587539.1 |
ZNF404 |
zinc finger protein 404 |
| chr1_-_85097431 | 0.22 |
ENST00000327308.3 |
C1orf180 |
chromosome 1 open reading frame 180 |
| chr1_+_61548225 | 0.22 |
ENST00000371187.3 |
NFIA |
nuclear factor I/A |
| chr19_+_41509851 | 0.22 |
ENST00000593831.1 ENST00000330446.5 |
CYP2B6 |
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr12_+_1929421 | 0.22 |
ENST00000543818.1 |
LRTM2 |
leucine-rich repeats and transmembrane domains 2 |
| chr10_-_79789291 | 0.22 |
ENST00000372371.3 |
POLR3A |
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr1_-_169599314 | 0.22 |
ENST00000367786.2 ENST00000458599.2 ENST00000367795.2 ENST00000263686.6 |
SELP |
selectin P (granule membrane protein 140kDa, antigen CD62) |
| chr3_+_69134124 | 0.22 |
ENST00000478935.1 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr3_-_157251383 | 0.21 |
ENST00000487753.1 ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
| chr12_+_1929783 | 0.21 |
ENST00000535041.1 |
LRTM2 |
leucine-rich repeats and transmembrane domains 2 |
| chr7_+_79998864 | 0.21 |
ENST00000435819.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr7_+_134551583 | 0.21 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chrX_-_99665262 | 0.21 |
ENST00000373034.4 ENST00000255531.7 |
PCDH19 |
protocadherin 19 |
| chr4_-_178363581 | 0.21 |
ENST00000264595.2 |
AGA |
aspartylglucosaminidase |
| chr12_+_1929644 | 0.21 |
ENST00000299194.1 |
LRTM2 |
leucine-rich repeats and transmembrane domains 2 |
| chr12_-_11214893 | 0.20 |
ENST00000533467.1 |
TAS2R46 |
taste receptor, type 2, member 46 |
| chr9_-_35815013 | 0.20 |
ENST00000259667.5 |
HINT2 |
histidine triad nucleotide binding protein 2 |
| chr3_+_196366555 | 0.20 |
ENST00000328557.4 |
NRROS |
negative regulator of reactive oxygen species |
| chr10_-_65028817 | 0.20 |
ENST00000542921.1 |
JMJD1C |
jumonji domain containing 1C |
| chr1_+_36549676 | 0.20 |
ENST00000207457.3 |
TEKT2 |
tektin 2 (testicular) |
| chr1_+_207039154 | 0.20 |
ENST00000367096.3 ENST00000391930.2 |
IL20 |
interleukin 20 |
| chr8_+_81397876 | 0.20 |
ENST00000430430.1 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
| chr4_-_159094194 | 0.20 |
ENST00000592057.1 ENST00000585682.1 ENST00000393807.5 |
FAM198B |
family with sequence similarity 198, member B |
| chrX_-_119763835 | 0.19 |
ENST00000371313.2 ENST00000304661.5 |
C1GALT1C1 |
C1GALT1-specific chaperone 1 |
| chr10_+_104535994 | 0.19 |
ENST00000369889.4 |
WBP1L |
WW domain binding protein 1-like |
| chr17_-_49124230 | 0.19 |
ENST00000510283.1 ENST00000510855.1 |
SPAG9 |
sperm associated antigen 9 |
| chr3_-_178984759 | 0.19 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr4_-_41750922 | 0.19 |
ENST00000226382.2 |
PHOX2B |
paired-like homeobox 2b |
| chr15_-_43785274 | 0.19 |
ENST00000413546.1 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr5_-_54468974 | 0.19 |
ENST00000381375.2 ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B |
cell division cycle 20B |
| chr9_+_124062071 | 0.19 |
ENST00000373818.4 |
GSN |
gelsolin |
| chr10_+_13142075 | 0.18 |
ENST00000378757.2 ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN |
optineurin |
| chr12_-_58027138 | 0.18 |
ENST00000341156.4 |
B4GALNT1 |
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr12_-_11287243 | 0.18 |
ENST00000539585.1 |
TAS2R30 |
taste receptor, type 2, member 30 |
| chr1_-_182369751 | 0.18 |
ENST00000367565.1 |
TEDDM1 |
transmembrane epididymal protein 1 |
| chr3_-_136471204 | 0.18 |
ENST00000480733.1 ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1 |
stromal antigen 1 |
| chr12_+_8309630 | 0.18 |
ENST00000396570.3 |
ZNF705A |
zinc finger protein 705A |
| chr9_-_215744 | 0.18 |
ENST00000382387.2 |
C9orf66 |
chromosome 9 open reading frame 66 |
| chr3_-_178789220 | 0.18 |
ENST00000414084.1 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr14_+_58894706 | 0.18 |
ENST00000261244.5 |
KIAA0586 |
KIAA0586 |
| chrX_-_13835147 | 0.17 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr16_-_18887627 | 0.17 |
ENST00000563235.1 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr4_+_26585686 | 0.17 |
ENST00000505206.1 ENST00000511789.1 |
TBC1D19 |
TBC1 domain family, member 19 |
| chrX_-_19905703 | 0.17 |
ENST00000397821.3 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr11_-_2182388 | 0.17 |
ENST00000421783.1 ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS INS-IGF2 |
insulin INS-IGF2 readthrough |
| chr4_-_16085340 | 0.17 |
ENST00000508167.1 |
PROM1 |
prominin 1 |
| chr10_-_65028938 | 0.17 |
ENST00000402544.1 |
JMJD1C |
jumonji domain containing 1C |
| chr9_+_35161998 | 0.17 |
ENST00000396787.1 ENST00000378495.3 ENST00000378496.4 |
UNC13B |
unc-13 homolog B (C. elegans) |
| chr5_+_135496675 | 0.16 |
ENST00000507637.1 |
SMAD5 |
SMAD family member 5 |
| chr2_+_201390843 | 0.16 |
ENST00000357799.4 ENST00000409203.3 |
SGOL2 |
shugoshin-like 2 (S. pombe) |
| chr17_+_79495397 | 0.16 |
ENST00000417245.2 ENST00000334850.7 |
FSCN2 |
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr8_-_57123815 | 0.16 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr4_-_16085314 | 0.16 |
ENST00000510224.1 |
PROM1 |
prominin 1 |
| chr3_+_63897605 | 0.16 |
ENST00000487717.1 |
ATXN7 |
ataxin 7 |
| chr7_+_28725585 | 0.16 |
ENST00000396298.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr3_+_88188254 | 0.15 |
ENST00000309495.5 |
ZNF654 |
zinc finger protein 654 |
| chr1_+_11866207 | 0.15 |
ENST00000312413.6 ENST00000346436.6 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr6_-_128841503 | 0.15 |
ENST00000368215.3 ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
| chr15_-_35261996 | 0.15 |
ENST00000156471.5 |
AQR |
aquarius intron-binding spliceosomal factor |
| chr12_-_109915098 | 0.14 |
ENST00000542858.1 ENST00000542262.1 ENST00000424763.2 |
KCTD10 |
potassium channel tetramerization domain containing 10 |
| chr3_+_152017360 | 0.14 |
ENST00000485910.1 ENST00000463374.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chrX_+_102611373 | 0.14 |
ENST00000372661.3 ENST00000372656.3 |
WBP5 |
WW domain binding protein 5 |
| chr16_-_21436459 | 0.14 |
ENST00000448012.2 ENST00000504841.2 ENST00000419180.2 |
NPIPB3 |
nuclear pore complex interacting protein family, member B3 |
| chr8_-_124553437 | 0.14 |
ENST00000517956.1 ENST00000443022.2 |
FBXO32 |
F-box protein 32 |
| chr17_-_35969409 | 0.14 |
ENST00000394378.2 ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG |
synergin, gamma |
| chr12_-_59314246 | 0.14 |
ENST00000320743.3 |
LRIG3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr1_+_218519577 | 0.14 |
ENST00000366930.4 ENST00000366929.4 |
TGFB2 |
transforming growth factor, beta 2 |
| chr8_+_66955648 | 0.13 |
ENST00000522619.1 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr16_-_21868978 | 0.13 |
ENST00000357370.5 ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chr11_+_118754475 | 0.13 |
ENST00000292174.4 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
| chr9_-_4666421 | 0.13 |
ENST00000381895.5 |
SPATA6L |
spermatogenesis associated 6-like |
| chr16_-_29415350 | 0.13 |
ENST00000524087.1 |
NPIPB11 |
nuclear pore complex interacting protein family, member B11 |
| chr12_-_11150474 | 0.13 |
ENST00000538986.1 |
TAS2R20 |
taste receptor, type 2, member 20 |
| chr3_-_71353892 | 0.13 |
ENST00000484350.1 |
FOXP1 |
forkhead box P1 |
| chr16_+_1583567 | 0.13 |
ENST00000566264.1 |
TMEM204 |
transmembrane protein 204 |
| chr6_+_10528560 | 0.13 |
ENST00000379597.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr5_+_49962772 | 0.13 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr14_+_75761099 | 0.13 |
ENST00000561000.1 ENST00000558575.1 |
RP11-293M10.5 |
RP11-293M10.5 |
| chr11_-_68780824 | 0.13 |
ENST00000441623.1 ENST00000309099.6 |
MRGPRF |
MAS-related GPR, member F |
| chr9_-_14314518 | 0.13 |
ENST00000397581.2 |
NFIB |
nuclear factor I/B |
| chr19_+_56152262 | 0.13 |
ENST00000325333.5 ENST00000590190.1 |
ZNF580 |
zinc finger protein 580 |
| chr1_+_61547405 | 0.12 |
ENST00000371189.4 |
NFIA |
nuclear factor I/A |
| chr15_+_23255242 | 0.12 |
ENST00000450802.3 |
GOLGA8I |
golgin A8 family, member I |
| chr17_-_39216344 | 0.12 |
ENST00000391418.2 |
KRTAP2-3 |
keratin associated protein 2-3 |
| chr15_-_43785303 | 0.12 |
ENST00000382039.3 ENST00000450115.2 ENST00000382044.4 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr12_+_104359641 | 0.12 |
ENST00000537100.1 |
TDG |
thymine-DNA glycosylase |
| chr17_+_56232494 | 0.12 |
ENST00000268912.5 |
OR4D1 |
olfactory receptor, family 4, subfamily D, member 1 |
| chr1_-_154164534 | 0.12 |
ENST00000271850.7 ENST00000368530.2 |
TPM3 |
tropomyosin 3 |
| chr12_-_7848364 | 0.12 |
ENST00000329913.3 |
GDF3 |
growth differentiation factor 3 |
| chr19_-_18392422 | 0.12 |
ENST00000252818.3 |
JUND |
jun D proto-oncogene |
| chr15_-_30685563 | 0.12 |
ENST00000401522.3 |
CHRFAM7A |
CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion |
| chr12_+_104359614 | 0.12 |
ENST00000266775.9 ENST00000544861.1 |
TDG |
thymine-DNA glycosylase |
| chr8_+_81397846 | 0.11 |
ENST00000379091.4 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
| chr6_+_41021027 | 0.11 |
ENST00000244669.2 |
APOBEC2 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr3_-_65583561 | 0.11 |
ENST00000460329.2 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr16_-_69418649 | 0.11 |
ENST00000566257.1 |
TERF2 |
telomeric repeat binding factor 2 |
| chr3_-_156878540 | 0.11 |
ENST00000461804.1 |
CCNL1 |
cyclin L1 |
| chr4_-_155471528 | 0.11 |
ENST00000302078.5 ENST00000499023.2 |
PLRG1 |
pleiotropic regulator 1 |
| chr1_+_367640 | 0.11 |
ENST00000426406.1 |
OR4F29 |
olfactory receptor, family 4, subfamily F, member 29 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.4 | 1.2 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 | 4.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 2.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.8 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 1.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 1.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.2 | 0.8 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.4 | GO:1902871 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.4 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.7 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.4 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.4 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.3 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.3 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.3 | GO:0050720 | interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.1 | 0.3 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.1 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.3 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.2 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 0.3 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 1.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.4 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.2 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.1 | 1.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.1 | 0.5 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.2 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.1 | 0.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.2 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 0.5 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.1 | 0.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.7 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.4 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.2 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.3 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:1903824 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.4 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.2 | GO:0061141 | lung ciliated cell differentiation(GO:0061141) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.2 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.3 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:2000566 | positive regulation of thymocyte migration(GO:2000412) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.1 | GO:2000439 | negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.5 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0006704 | androgen biosynthetic process(GO:0006702) glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) renal water absorption(GO:0070295) |
| 0.0 | 0.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 1.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.0 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.2 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.0 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 9.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 4.6 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 2.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.3 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 1.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.4 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 1.6 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.0 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.0 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 2.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.6 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.1 | 0.4 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 6.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.2 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.1 | 1.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.4 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.5 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.2 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.1 | 1.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 9.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.3 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 1.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.9 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.0 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 9.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |