Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for NFIA
Z-value: 0.24
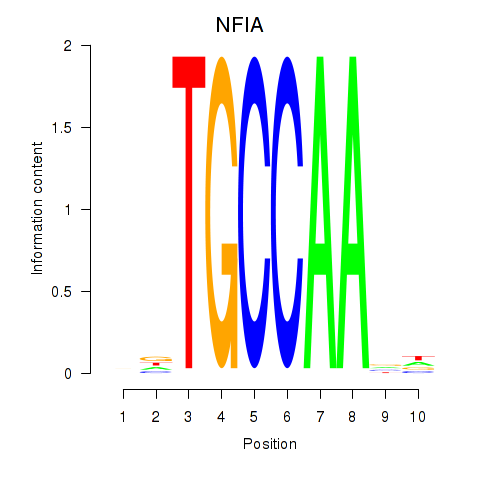
Transcription factors associated with NFIA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFIA
|
ENSG00000162599.11 | NFIA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFIA | hg19_v2_chr1_+_61547894_61547980 | -0.34 | 4.2e-01 | Click! |
Activity profile of NFIA motif
Sorted Z-values of NFIA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFIA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_158345462 | 0.50 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr1_-_19186176 | 0.49 |
ENST00000375371.3 |
TAS1R2 |
taste receptor, type 1, member 2 |
| chr4_+_30721968 | 0.41 |
ENST00000361762.2 |
PCDH7 |
protocadherin 7 |
| chr2_-_111291587 | 0.37 |
ENST00000437167.1 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
| chr11_+_10326612 | 0.37 |
ENST00000534464.1 ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM |
adrenomedullin |
| chr2_-_163100045 | 0.37 |
ENST00000188790.4 |
FAP |
fibroblast activation protein, alpha |
| chr2_-_163099885 | 0.36 |
ENST00000443424.1 |
FAP |
fibroblast activation protein, alpha |
| chr12_-_52946923 | 0.35 |
ENST00000267119.5 |
KRT71 |
keratin 71 |
| chr4_+_113739244 | 0.34 |
ENST00000503271.1 ENST00000503423.1 ENST00000506722.1 |
ANK2 |
ankyrin 2, neuronal |
| chr4_+_113970772 | 0.33 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr7_-_141646726 | 0.32 |
ENST00000438351.1 ENST00000439991.1 ENST00000551012.2 ENST00000546910.1 |
CLEC5A |
C-type lectin domain family 5, member A |
| chr12_-_10251539 | 0.30 |
ENST00000420265.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr17_-_15168624 | 0.30 |
ENST00000312280.3 ENST00000494511.1 ENST00000580584.1 |
PMP22 |
peripheral myelin protein 22 |
| chr2_-_40739501 | 0.29 |
ENST00000403092.1 ENST00000402441.1 ENST00000448531.1 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr17_-_39538550 | 0.27 |
ENST00000394001.1 |
KRT34 |
keratin 34 |
| chr12_-_7281469 | 0.27 |
ENST00000542370.1 ENST00000266560.3 |
RBP5 |
retinol binding protein 5, cellular |
| chr17_+_1633755 | 0.27 |
ENST00000545662.1 |
WDR81 |
WD repeat domain 81 |
| chr7_+_120629653 | 0.26 |
ENST00000450913.2 ENST00000340646.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr7_-_19184929 | 0.26 |
ENST00000275461.3 |
FERD3L |
Fer3-like bHLH transcription factor |
| chr11_-_2170786 | 0.25 |
ENST00000300632.5 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr8_+_97597148 | 0.25 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr19_+_41725140 | 0.25 |
ENST00000359092.3 |
AXL |
AXL receptor tyrosine kinase |
| chr5_+_112074029 | 0.25 |
ENST00000512211.2 |
APC |
adenomatous polyposis coli |
| chr8_+_32579341 | 0.24 |
ENST00000519240.1 ENST00000539990.1 |
NRG1 |
neuregulin 1 |
| chr11_-_35547151 | 0.24 |
ENST00000378878.3 ENST00000529303.1 ENST00000278360.3 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr2_+_189839046 | 0.24 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr19_+_41725088 | 0.23 |
ENST00000301178.4 |
AXL |
AXL receptor tyrosine kinase |
| chr11_+_15136462 | 0.23 |
ENST00000379556.3 ENST00000424273.1 |
INSC |
inscuteable homolog (Drosophila) |
| chr12_-_71003568 | 0.22 |
ENST00000547715.1 ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr2_-_145277569 | 0.22 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr1_+_95975672 | 0.21 |
ENST00000440116.2 ENST00000456933.1 |
RP11-286B14.1 |
RP11-286B14.1 |
| chr17_+_54671047 | 0.21 |
ENST00000332822.4 |
NOG |
noggin |
| chr14_+_23842018 | 0.21 |
ENST00000397242.2 ENST00000329715.2 |
IL25 |
interleukin 25 |
| chrX_-_69479654 | 0.21 |
ENST00000374519.2 |
P2RY4 |
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr19_-_12997995 | 0.21 |
ENST00000264834.4 |
KLF1 |
Kruppel-like factor 1 (erythroid) |
| chr5_+_112073544 | 0.21 |
ENST00000257430.4 ENST00000508376.2 |
APC |
adenomatous polyposis coli |
| chr4_-_157892055 | 0.21 |
ENST00000422544.2 |
PDGFC |
platelet derived growth factor C |
| chr8_+_104384616 | 0.20 |
ENST00000520337.1 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr17_-_8113542 | 0.20 |
ENST00000578549.1 ENST00000535053.1 ENST00000582368.1 |
AURKB |
aurora kinase B |
| chr11_-_104480019 | 0.20 |
ENST00000536529.1 ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1 |
RP11-886D15.1 |
| chrX_+_49832231 | 0.19 |
ENST00000376108.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr9_+_130911770 | 0.19 |
ENST00000372998.1 |
LCN2 |
lipocalin 2 |
| chr8_+_38586068 | 0.19 |
ENST00000443286.2 ENST00000520340.1 ENST00000518415.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr1_+_107683436 | 0.19 |
ENST00000370068.1 |
NTNG1 |
netrin G1 |
| chr10_-_106240032 | 0.18 |
ENST00000447860.1 |
RP11-127O4.3 |
RP11-127O4.3 |
| chr9_+_130911723 | 0.18 |
ENST00000277480.2 ENST00000373013.2 ENST00000540948.1 |
LCN2 |
lipocalin 2 |
| chr1_+_34632484 | 0.18 |
ENST00000373374.3 |
C1orf94 |
chromosome 1 open reading frame 94 |
| chr3_-_105588231 | 0.17 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr6_+_106959718 | 0.17 |
ENST00000369066.3 |
AIM1 |
absent in melanoma 1 |
| chr8_-_131399110 | 0.17 |
ENST00000521426.1 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr5_+_50678921 | 0.17 |
ENST00000230658.7 |
ISL1 |
ISL LIM homeobox 1 |
| chr12_-_13248598 | 0.17 |
ENST00000337630.6 ENST00000545699.1 |
GSG1 |
germ cell associated 1 |
| chr2_+_30569506 | 0.17 |
ENST00000421976.2 |
AC109642.1 |
AC109642.1 |
| chrX_-_83757399 | 0.17 |
ENST00000373177.2 ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX |
highly divergent homeobox |
| chr16_-_10652993 | 0.17 |
ENST00000536829.1 |
EMP2 |
epithelial membrane protein 2 |
| chr3_+_148447887 | 0.17 |
ENST00000475347.1 ENST00000474935.1 ENST00000461609.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr5_+_53751445 | 0.17 |
ENST00000302005.1 |
HSPB3 |
heat shock 27kDa protein 3 |
| chrX_+_105192423 | 0.17 |
ENST00000540278.1 |
NRK |
Nik related kinase |
| chr16_+_31404624 | 0.17 |
ENST00000389202.2 |
ITGAD |
integrin, alpha D |
| chr1_+_107683644 | 0.17 |
ENST00000370067.1 |
NTNG1 |
netrin G1 |
| chr11_+_118230287 | 0.16 |
ENST00000252108.3 ENST00000431736.2 |
UBE4A |
ubiquitination factor E4A |
| chr14_-_94984181 | 0.16 |
ENST00000341228.2 |
SERPINA12 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr1_-_114429997 | 0.16 |
ENST00000471267.1 ENST00000393320.3 |
BCL2L15 |
BCL2-like 15 |
| chr13_+_102104952 | 0.15 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr16_+_82090028 | 0.15 |
ENST00000568090.1 |
HSD17B2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr1_+_22962948 | 0.15 |
ENST00000374642.3 |
C1QA |
complement component 1, q subcomponent, A chain |
| chr8_+_38585704 | 0.15 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr5_-_64777733 | 0.15 |
ENST00000381055.3 |
ADAMTS6 |
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr13_+_102104980 | 0.15 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr1_+_160160283 | 0.15 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chrX_-_49965009 | 0.15 |
ENST00000437370.1 ENST00000376064.3 ENST00000448865.1 |
AKAP4 |
A kinase (PRKA) anchor protein 4 |
| chr17_-_8702667 | 0.14 |
ENST00000329805.4 |
MFSD6L |
major facilitator superfamily domain containing 6-like |
| chr6_+_123100620 | 0.14 |
ENST00000368444.3 |
FABP7 |
fatty acid binding protein 7, brain |
| chr1_+_160160346 | 0.14 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chrX_+_153770421 | 0.14 |
ENST00000369609.5 ENST00000369607.1 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr2_+_152214098 | 0.14 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr7_-_83824169 | 0.14 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr9_-_130541017 | 0.14 |
ENST00000314830.8 |
SH2D3C |
SH2 domain containing 3C |
| chrY_+_26997726 | 0.14 |
ENST00000382296.2 |
DAZ4 |
deleted in azoospermia 4 |
| chr9_+_37667978 | 0.14 |
ENST00000539465.1 |
FRMPD1 |
FERM and PDZ domain containing 1 |
| chr17_-_39661849 | 0.14 |
ENST00000246635.3 ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13 |
keratin 13 |
| chr2_-_190044480 | 0.13 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr10_-_104597286 | 0.13 |
ENST00000369887.3 |
CYP17A1 |
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr4_-_114900831 | 0.13 |
ENST00000315366.7 |
ARSJ |
arylsulfatase family, member J |
| chr1_+_32084794 | 0.13 |
ENST00000373705.1 |
HCRTR1 |
hypocretin (orexin) receptor 1 |
| chr14_-_61124977 | 0.13 |
ENST00000554986.1 |
SIX1 |
SIX homeobox 1 |
| chr19_-_33462816 | 0.13 |
ENST00000305768.5 ENST00000590597.2 |
CEP89 |
centrosomal protein 89kDa |
| chr5_+_135394840 | 0.13 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chrX_-_19817869 | 0.13 |
ENST00000379698.4 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr17_+_16120512 | 0.13 |
ENST00000581006.1 ENST00000584797.1 ENST00000498772.2 ENST00000225609.5 ENST00000395844.4 ENST00000477745.1 |
PIGL |
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr10_+_5238793 | 0.12 |
ENST00000263126.1 |
AKR1C4 |
aldo-keto reductase family 1, member C4 |
| chr6_-_30658745 | 0.12 |
ENST00000376420.5 ENST00000376421.5 |
NRM |
nurim (nuclear envelope membrane protein) |
| chr4_-_113437328 | 0.12 |
ENST00000313341.3 |
NEUROG2 |
neurogenin 2 |
| chr12_-_53045948 | 0.12 |
ENST00000309680.3 |
KRT2 |
keratin 2 |
| chr7_+_100199800 | 0.12 |
ENST00000223061.5 |
PCOLCE |
procollagen C-endopeptidase enhancer |
| chr19_-_13227534 | 0.12 |
ENST00000588229.1 ENST00000357720.4 |
TRMT1 |
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr10_-_24770632 | 0.12 |
ENST00000596413.1 |
AL353583.1 |
AL353583.1 |
| chr14_-_85996332 | 0.12 |
ENST00000380722.1 |
RP11-497E19.1 |
RP11-497E19.1 |
| chr19_-_40440533 | 0.12 |
ENST00000221347.6 |
FCGBP |
Fc fragment of IgG binding protein |
| chr12_+_122064398 | 0.12 |
ENST00000330079.7 |
ORAI1 |
ORAI calcium release-activated calcium modulator 1 |
| chr1_+_170501270 | 0.12 |
ENST00000367763.3 ENST00000367762.1 |
GORAB |
golgin, RAB6-interacting |
| chr19_-_13227463 | 0.12 |
ENST00000437766.1 ENST00000221504.8 |
TRMT1 |
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr13_+_78109804 | 0.12 |
ENST00000535157.1 |
SCEL |
sciellin |
| chrX_-_15332665 | 0.12 |
ENST00000537676.1 ENST00000344384.4 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr12_+_113229543 | 0.12 |
ENST00000447659.2 |
RPH3A |
rabphilin 3A homolog (mouse) |
| chr12_+_113229737 | 0.12 |
ENST00000551052.1 ENST00000415485.3 |
RPH3A |
rabphilin 3A homolog (mouse) |
| chr9_-_93405352 | 0.11 |
ENST00000375765.3 |
DIRAS2 |
DIRAS family, GTP-binding RAS-like 2 |
| chr4_-_157892167 | 0.11 |
ENST00000541126.1 |
PDGFC |
platelet derived growth factor C |
| chr18_-_31802282 | 0.11 |
ENST00000535475.1 |
NOL4 |
nucleolar protein 4 |
| chr18_+_28956740 | 0.11 |
ENST00000308128.4 ENST00000359747.4 |
DSG4 |
desmoglein 4 |
| chr1_+_32930647 | 0.11 |
ENST00000609129.1 |
ZBTB8B |
zinc finger and BTB domain containing 8B |
| chr3_+_184033135 | 0.11 |
ENST00000424196.1 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_+_214163033 | 0.11 |
ENST00000607425.1 |
PROX1 |
prospero homeobox 1 |
| chr18_-_31802056 | 0.11 |
ENST00000538587.1 |
NOL4 |
nucleolar protein 4 |
| chr1_+_78769549 | 0.11 |
ENST00000370758.1 |
PTGFR |
prostaglandin F receptor (FP) |
| chr13_+_113622757 | 0.11 |
ENST00000375604.2 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr1_+_32084641 | 0.11 |
ENST00000373706.5 |
HCRTR1 |
hypocretin (orexin) receptor 1 |
| chr10_+_115312766 | 0.11 |
ENST00000351270.3 |
HABP2 |
hyaluronan binding protein 2 |
| chr3_+_97887544 | 0.11 |
ENST00000356526.2 |
OR5H15 |
olfactory receptor, family 5, subfamily H, member 15 |
| chr6_+_107077471 | 0.11 |
ENST00000369044.1 |
QRSL1 |
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr3_+_101498074 | 0.11 |
ENST00000273347.5 ENST00000474165.1 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
| chr3_-_121740969 | 0.11 |
ENST00000393631.1 ENST00000273691.3 ENST00000344209.5 |
ILDR1 |
immunoglobulin-like domain containing receptor 1 |
| chr7_+_128312346 | 0.11 |
ENST00000480462.1 ENST00000378704.3 ENST00000477515.1 |
FAM71F2 |
family with sequence similarity 71, member F2 |
| chr13_+_78109884 | 0.11 |
ENST00000377246.3 ENST00000349847.3 |
SCEL |
sciellin |
| chr2_+_241938255 | 0.11 |
ENST00000401884.1 ENST00000405547.3 ENST00000310397.8 ENST00000342631.6 |
SNED1 |
sushi, nidogen and EGF-like domains 1 |
| chr19_-_41196458 | 0.11 |
ENST00000598779.1 |
NUMBL |
numb homolog (Drosophila)-like |
| chr17_-_53809473 | 0.11 |
ENST00000575734.1 |
TMEM100 |
transmembrane protein 100 |
| chr11_+_193065 | 0.11 |
ENST00000342878.2 |
SCGB1C1 |
secretoglobin, family 1C, member 1 |
| chr6_-_122792919 | 0.10 |
ENST00000339697.4 |
SERINC1 |
serine incorporator 1 |
| chrX_+_105855160 | 0.10 |
ENST00000372544.2 ENST00000372548.4 |
CXorf57 |
chromosome X open reading frame 57 |
| chr5_-_141249154 | 0.10 |
ENST00000357517.5 ENST00000536585.1 |
PCDH1 |
protocadherin 1 |
| chr11_+_20044096 | 0.10 |
ENST00000533917.1 |
NAV2 |
neuron navigator 2 |
| chr17_+_7323634 | 0.10 |
ENST00000323675.3 |
SPEM1 |
spermatid maturation 1 |
| chr5_+_43602750 | 0.10 |
ENST00000505678.2 ENST00000512422.1 ENST00000264663.5 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr18_+_61144160 | 0.10 |
ENST00000489441.1 ENST00000424602.1 |
SERPINB5 |
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr12_-_11150474 | 0.10 |
ENST00000538986.1 |
TAS2R20 |
taste receptor, type 2, member 20 |
| chr3_+_184032919 | 0.10 |
ENST00000427845.1 ENST00000342981.4 ENST00000319274.6 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr3_-_123411191 | 0.10 |
ENST00000354792.5 ENST00000508240.1 |
MYLK |
myosin light chain kinase |
| chr12_-_66563786 | 0.10 |
ENST00000542724.1 |
TMBIM4 |
transmembrane BAX inhibitor motif containing 4 |
| chr1_-_203144941 | 0.10 |
ENST00000255416.4 |
MYBPH |
myosin binding protein H |
| chr19_-_13947099 | 0.10 |
ENST00000587762.1 |
MIR24-2 |
microRNA 24-2 |
| chr18_-_5396271 | 0.10 |
ENST00000579951.1 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr3_-_155011483 | 0.10 |
ENST00000489090.1 |
RP11-451G4.2 |
RP11-451G4.2 |
| chr9_+_71944241 | 0.10 |
ENST00000257515.8 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr15_+_67458357 | 0.10 |
ENST00000537194.2 |
SMAD3 |
SMAD family member 3 |
| chr1_-_26394114 | 0.10 |
ENST00000374272.3 |
TRIM63 |
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr12_-_6716569 | 0.10 |
ENST00000544040.1 ENST00000545942.1 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
| chr19_-_46296011 | 0.10 |
ENST00000377735.3 ENST00000270223.6 |
DMWD |
dystrophia myotonica, WD repeat containing |
| chr16_-_55866997 | 0.09 |
ENST00000360526.3 ENST00000361503.4 |
CES1 |
carboxylesterase 1 |
| chr16_-_15474904 | 0.09 |
ENST00000534094.1 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
| chr2_-_231989808 | 0.09 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr8_-_16859690 | 0.09 |
ENST00000180166.5 |
FGF20 |
fibroblast growth factor 20 |
| chr11_-_5271122 | 0.09 |
ENST00000330597.3 |
HBG1 |
hemoglobin, gamma A |
| chr5_-_142783694 | 0.09 |
ENST00000394466.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr11_-_58343319 | 0.09 |
ENST00000395074.2 |
LPXN |
leupaxin |
| chr12_-_131200810 | 0.09 |
ENST00000536002.1 ENST00000544034.1 |
RIMBP2 RP11-662M24.2 |
RIMS binding protein 2 RP11-662M24.2 |
| chr11_-_61348292 | 0.09 |
ENST00000539008.1 ENST00000540677.1 ENST00000542836.1 ENST00000542670.1 ENST00000535826.1 ENST00000545053.1 |
SYT7 |
synaptotagmin VII |
| chr14_-_50319758 | 0.09 |
ENST00000298310.5 |
NEMF |
nuclear export mediator factor |
| chr4_+_14113592 | 0.09 |
ENST00000502759.1 ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085 |
long intergenic non-protein coding RNA 1085 |
| chr2_+_172864490 | 0.09 |
ENST00000315796.4 |
METAP1D |
methionyl aminopeptidase type 1D (mitochondrial) |
| chr7_+_62809239 | 0.09 |
ENST00000456890.1 |
AC006455.1 |
AC006455.1 |
| chr16_-_55867146 | 0.09 |
ENST00000422046.2 |
CES1 |
carboxylesterase 1 |
| chr11_-_122931881 | 0.09 |
ENST00000526110.1 ENST00000227378.3 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr14_-_98444438 | 0.09 |
ENST00000512901.2 |
C14orf64 |
chromosome 14 open reading frame 64 |
| chr6_-_9933500 | 0.09 |
ENST00000492169.1 |
OFCC1 |
orofacial cleft 1 candidate 1 |
| chr13_-_46716969 | 0.09 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr11_-_113644491 | 0.08 |
ENST00000200135.3 |
ZW10 |
zw10 kinetochore protein |
| chr5_-_43313574 | 0.08 |
ENST00000325110.6 ENST00000433297.2 |
HMGCS1 |
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr4_-_21699380 | 0.08 |
ENST00000382148.3 |
KCNIP4 |
Kv channel interacting protein 4 |
| chr13_-_29069232 | 0.08 |
ENST00000282397.4 ENST00000541932.1 ENST00000539099.1 |
FLT1 |
fms-related tyrosine kinase 1 |
| chr17_+_35732916 | 0.08 |
ENST00000586700.1 |
C17orf78 |
chromosome 17 open reading frame 78 |
| chr11_-_26593677 | 0.08 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr8_+_42873548 | 0.08 |
ENST00000533338.1 ENST00000534420.1 |
HOOK3 RP11-598P20.5 |
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr1_-_1009683 | 0.08 |
ENST00000453464.2 |
RNF223 |
ring finger protein 223 |
| chr17_+_35732955 | 0.08 |
ENST00000300618.4 |
C17orf78 |
chromosome 17 open reading frame 78 |
| chr16_-_19725899 | 0.08 |
ENST00000567367.1 |
KNOP1 |
lysine-rich nucleolar protein 1 |
| chr19_-_1812193 | 0.08 |
ENST00000525591.1 |
ATP8B3 |
ATPase, aminophospholipid transporter, class I, type 8B, member 3 |
| chr5_+_140868717 | 0.08 |
ENST00000252087.1 |
PCDHGC5 |
protocadherin gamma subfamily C, 5 |
| chr8_-_22089533 | 0.08 |
ENST00000321613.3 |
PHYHIP |
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr19_+_36249057 | 0.08 |
ENST00000301165.5 ENST00000536950.1 ENST00000537459.1 ENST00000421853.2 |
C19orf55 |
chromosome 19 open reading frame 55 |
| chrX_-_13835147 | 0.08 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr13_-_40177261 | 0.08 |
ENST00000379589.3 |
LHFP |
lipoma HMGIC fusion partner |
| chr11_-_16430399 | 0.08 |
ENST00000528252.1 |
SOX6 |
SRY (sex determining region Y)-box 6 |
| chr11_+_5423827 | 0.08 |
ENST00000332043.1 |
OR51J1 |
olfactory receptor, family 51, subfamily J, member 1 (gene/pseudogene) |
| chr4_-_76928641 | 0.08 |
ENST00000264888.5 |
CXCL9 |
chemokine (C-X-C motif) ligand 9 |
| chr17_-_1733114 | 0.08 |
ENST00000305513.7 |
SMYD4 |
SET and MYND domain containing 4 |
| chr1_-_243326612 | 0.08 |
ENST00000492145.1 ENST00000490813.1 ENST00000464936.1 |
CEP170 |
centrosomal protein 170kDa |
| chr7_+_17338239 | 0.08 |
ENST00000242057.4 |
AHR |
aryl hydrocarbon receptor |
| chr17_-_39093672 | 0.08 |
ENST00000209718.3 ENST00000436344.3 ENST00000485751.1 |
KRT23 |
keratin 23 (histone deacetylase inducible) |
| chr2_-_176866978 | 0.08 |
ENST00000392540.2 ENST00000409660.1 ENST00000544803.1 ENST00000272748.4 |
KIAA1715 |
KIAA1715 |
| chr9_+_17134980 | 0.07 |
ENST00000380647.3 |
CNTLN |
centlein, centrosomal protein |
| chr11_+_60997055 | 0.07 |
ENST00000544899.1 |
PGA4 |
pepsinogen 4, group I (pepsinogen A) |
| chr4_+_166248775 | 0.07 |
ENST00000261507.6 ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1 |
methylsterol monooxygenase 1 |
| chr3_-_137851220 | 0.07 |
ENST00000236709.3 |
A4GNT |
alpha-1,4-N-acetylglucosaminyltransferase |
| chr19_-_41196534 | 0.07 |
ENST00000252891.4 |
NUMBL |
numb homolog (Drosophila)-like |
| chr10_-_61122220 | 0.07 |
ENST00000422313.2 ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C |
family with sequence similarity 13, member C |
| chr11_-_26593779 | 0.07 |
ENST00000529533.1 |
MUC15 |
mucin 15, cell surface associated |
| chr12_+_101188718 | 0.07 |
ENST00000299222.9 ENST00000392977.3 |
ANO4 |
anoctamin 4 |
| chr4_-_103266355 | 0.07 |
ENST00000424970.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr7_-_99381884 | 0.07 |
ENST00000336411.2 |
CYP3A4 |
cytochrome P450, family 3, subfamily A, polypeptide 4 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 0.7 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 0.5 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.6 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.4 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.3 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.1 | 0.3 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 0.2 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.2 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.1 | 0.4 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 0.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.4 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:0036378 | alkaloid catabolic process(GO:0009822) calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.3 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.2 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.2 | GO:0009624 | response to nematode(GO:0009624) eosinophil differentiation(GO:0030222) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.5 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.1 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.0 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.3 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.0 | GO:0060459 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.0 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.5 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.1 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.2 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.6 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |