Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
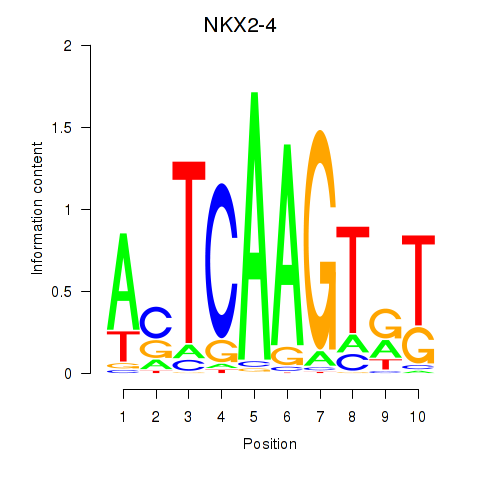
Results for NKX2-4
Z-value: 0.59
Transcription factors associated with NKX2-4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-4
|
ENSG00000125816.4 | NKX2-4 |
Activity profile of NKX2-4 motif
Sorted Z-values of NKX2-4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_97597148 | 0.82 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr17_-_53800217 | 0.47 |
ENST00000424486.2 |
TMEM100 |
transmembrane protein 100 |
| chr4_-_100242549 | 0.40 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr2_-_188419200 | 0.36 |
ENST00000233156.3 ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr1_-_85870177 | 0.32 |
ENST00000542148.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr2_-_188419078 | 0.32 |
ENST00000437725.1 ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr9_-_95244781 | 0.31 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr17_-_67138015 | 0.31 |
ENST00000284425.2 ENST00000590645.1 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr4_-_175443943 | 0.31 |
ENST00000296522.6 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr9_+_90112767 | 0.30 |
ENST00000408954.3 |
DAPK1 |
death-associated protein kinase 1 |
| chr9_-_95166841 | 0.30 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr9_+_90112741 | 0.29 |
ENST00000469640.2 |
DAPK1 |
death-associated protein kinase 1 |
| chr6_+_101846664 | 0.29 |
ENST00000421544.1 ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
| chr7_-_47579188 | 0.29 |
ENST00000398879.1 ENST00000355730.3 ENST00000442536.2 ENST00000458317.2 |
TNS3 |
tensin 3 |
| chr9_-_95166884 | 0.29 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr1_+_64239657 | 0.28 |
ENST00000371080.1 ENST00000371079.1 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
| chr6_-_152958521 | 0.26 |
ENST00000367255.5 ENST00000265368.4 ENST00000448038.1 ENST00000341594.5 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr7_+_129932974 | 0.25 |
ENST00000445470.2 ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4 |
carboxypeptidase A4 |
| chr4_+_86749045 | 0.24 |
ENST00000514229.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr10_-_28571015 | 0.23 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_+_110036728 | 0.23 |
ENST00000369868.3 ENST00000430195.2 |
CYB561D1 |
cytochrome b561 family, member D1 |
| chr1_+_110036699 | 0.23 |
ENST00000496961.1 ENST00000533024.1 ENST00000310611.4 ENST00000527072.1 ENST00000420578.2 ENST00000528785.1 |
CYB561D1 |
cytochrome b561 family, member D1 |
| chr1_-_76398077 | 0.23 |
ENST00000284142.6 |
ASB17 |
ankyrin repeat and SOCS box containing 17 |
| chr1_+_26146397 | 0.22 |
ENST00000374303.2 ENST00000533762.1 ENST00000529116.1 ENST00000474295.1 ENST00000488327.2 ENST00000472643.1 ENST00000526894.1 ENST00000524618.1 ENST00000374307.5 |
MTFR1L |
mitochondrial fission regulator 1-like |
| chr1_-_12891264 | 0.22 |
ENST00000535591.1 ENST00000437584.1 |
PRAMEF11 |
PRAME family member 11 |
| chr6_-_152639479 | 0.21 |
ENST00000356820.4 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr5_+_137419581 | 0.21 |
ENST00000506684.1 ENST00000504809.1 ENST00000398754.1 |
WNT8A |
wingless-type MMTV integration site family, member 8A |
| chr7_+_120629653 | 0.21 |
ENST00000450913.2 ENST00000340646.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr18_+_66465302 | 0.21 |
ENST00000360242.5 ENST00000358653.5 |
CCDC102B |
coiled-coil domain containing 102B |
| chr21_+_17553910 | 0.21 |
ENST00000428669.2 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr15_-_82338460 | 0.21 |
ENST00000558133.1 ENST00000329713.4 |
MEX3B |
mex-3 RNA binding family member B |
| chr7_-_72739575 | 0.21 |
ENST00000453152.1 |
TRIM50 |
tripartite motif containing 50 |
| chr1_+_226013047 | 0.20 |
ENST00000366837.4 |
EPHX1 |
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr3_-_112329110 | 0.19 |
ENST00000479368.1 |
CCDC80 |
coiled-coil domain containing 80 |
| chr1_+_152850787 | 0.19 |
ENST00000368765.3 |
SMCP |
sperm mitochondria-associated cysteine-rich protein |
| chr13_-_43566301 | 0.19 |
ENST00000398762.3 ENST00000313640.7 ENST00000313624.7 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
| chr8_-_6914251 | 0.19 |
ENST00000330590.2 |
DEFA5 |
defensin, alpha 5, Paneth cell-specific |
| chr10_-_49482907 | 0.19 |
ENST00000374201.3 ENST00000407470.4 |
FRMPD2 |
FERM and PDZ domain containing 2 |
| chr5_-_160279207 | 0.18 |
ENST00000327245.5 |
ATP10B |
ATPase, class V, type 10B |
| chr11_-_82746587 | 0.18 |
ENST00000528379.1 ENST00000534103.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr17_-_15168624 | 0.17 |
ENST00000312280.3 ENST00000494511.1 ENST00000580584.1 |
PMP22 |
peripheral myelin protein 22 |
| chr11_-_102826434 | 0.17 |
ENST00000340273.4 ENST00000260302.3 |
MMP13 |
matrix metallopeptidase 13 (collagenase 3) |
| chr1_+_144989309 | 0.17 |
ENST00000596396.1 |
AL590452.1 |
Uncharacterized protein |
| chr11_-_105892937 | 0.17 |
ENST00000301919.4 ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4 |
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr1_-_204183071 | 0.16 |
ENST00000308302.3 |
GOLT1A |
golgi transport 1A |
| chr12_-_71182695 | 0.16 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr7_+_134551583 | 0.16 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr1_+_196788887 | 0.14 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr7_-_14028488 | 0.14 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr12_+_75874460 | 0.14 |
ENST00000266659.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr8_+_22429205 | 0.14 |
ENST00000520207.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr19_+_41117770 | 0.14 |
ENST00000601032.1 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr2_+_161993465 | 0.14 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr9_+_140119618 | 0.14 |
ENST00000359069.2 |
C9orf169 |
chromosome 9 open reading frame 169 |
| chr16_-_31161380 | 0.13 |
ENST00000569305.1 ENST00000418068.2 ENST00000268281.4 |
PRSS36 |
protease, serine, 36 |
| chr14_+_51955831 | 0.13 |
ENST00000356218.4 |
FRMD6 |
FERM domain containing 6 |
| chr9_-_95298314 | 0.13 |
ENST00000344604.5 ENST00000375540.1 |
ECM2 |
extracellular matrix protein 2, female organ and adipocyte specific |
| chr19_-_51845378 | 0.13 |
ENST00000335624.4 |
VSIG10L |
V-set and immunoglobulin domain containing 10 like |
| chr2_+_54951679 | 0.13 |
ENST00000356458.6 |
EML6 |
echinoderm microtubule associated protein like 6 |
| chr17_-_63822563 | 0.13 |
ENST00000317442.8 |
CEP112 |
centrosomal protein 112kDa |
| chr3_-_18480260 | 0.13 |
ENST00000454909.2 |
SATB1 |
SATB homeobox 1 |
| chr12_-_58329819 | 0.13 |
ENST00000551421.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chrX_+_102883620 | 0.13 |
ENST00000372626.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr2_-_175462934 | 0.12 |
ENST00000392546.2 ENST00000436221.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr9_+_37667978 | 0.12 |
ENST00000539465.1 |
FRMPD1 |
FERM and PDZ domain containing 1 |
| chr4_+_86748898 | 0.12 |
ENST00000509300.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr15_+_63334831 | 0.12 |
ENST00000288398.6 ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr11_-_104035088 | 0.12 |
ENST00000302251.5 |
PDGFD |
platelet derived growth factor D |
| chr7_+_151038850 | 0.12 |
ENST00000355851.4 ENST00000566856.1 ENST00000470229.1 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
| chr1_-_89591749 | 0.12 |
ENST00000370466.3 |
GBP2 |
guanylate binding protein 2, interferon-inducible |
| chr2_-_175462456 | 0.12 |
ENST00000409891.1 ENST00000410117.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr1_-_241520525 | 0.11 |
ENST00000366565.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr1_+_161632937 | 0.11 |
ENST00000236937.9 ENST00000367961.4 ENST00000358671.5 |
FCGR2B |
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr5_+_139505520 | 0.11 |
ENST00000333305.3 |
IGIP |
IgA-inducing protein |
| chr19_-_50316423 | 0.11 |
ENST00000528094.1 ENST00000526575.1 |
FUZ |
fuzzy planar cell polarity protein |
| chr3_+_137906109 | 0.11 |
ENST00000481646.1 ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr12_-_48500085 | 0.11 |
ENST00000549518.1 |
SENP1 |
SUMO1/sentrin specific peptidase 1 |
| chr9_+_124048864 | 0.11 |
ENST00000545652.1 |
GSN |
gelsolin |
| chr19_-_50316517 | 0.11 |
ENST00000313777.4 ENST00000445575.2 |
FUZ |
fuzzy planar cell polarity protein |
| chr2_+_161993412 | 0.11 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr6_+_45390222 | 0.11 |
ENST00000359524.5 |
RUNX2 |
runt-related transcription factor 2 |
| chr8_+_11141925 | 0.11 |
ENST00000221086.3 |
MTMR9 |
myotubularin related protein 9 |
| chr6_-_41703952 | 0.11 |
ENST00000358871.2 ENST00000403298.4 |
TFEB |
transcription factor EB |
| chr19_-_50316489 | 0.11 |
ENST00000533418.1 |
FUZ |
fuzzy planar cell polarity protein |
| chr11_+_35965531 | 0.11 |
ENST00000528989.1 ENST00000524419.1 ENST00000315571.5 |
LDLRAD3 |
low density lipoprotein receptor class A domain containing 3 |
| chr12_+_54422142 | 0.10 |
ENST00000243108.4 |
HOXC6 |
homeobox C6 |
| chr11_+_12766583 | 0.10 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr14_-_88459182 | 0.10 |
ENST00000544807.2 |
GALC |
galactosylceramidase |
| chr12_+_109826524 | 0.10 |
ENST00000431443.2 |
MYO1H |
myosin IH |
| chr15_-_20193370 | 0.10 |
ENST00000558565.2 |
IGHV3OR15-7 |
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr14_-_105708942 | 0.10 |
ENST00000549655.1 |
BRF1 |
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
| chr12_+_133757995 | 0.10 |
ENST00000536435.2 ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268 |
zinc finger protein 268 |
| chr1_+_202431859 | 0.10 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr5_+_147582348 | 0.10 |
ENST00000514389.1 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr16_+_3062457 | 0.10 |
ENST00000445369.2 |
CLDN9 |
claudin 9 |
| chr20_-_39928705 | 0.10 |
ENST00000436099.2 ENST00000309060.3 ENST00000373261.1 ENST00000436440.2 ENST00000540170.1 ENST00000557816.1 ENST00000560361.1 |
ZHX3 |
zinc fingers and homeoboxes 3 |
| chr8_+_22132847 | 0.10 |
ENST00000521356.1 |
PIWIL2 |
piwi-like RNA-mediated gene silencing 2 |
| chr2_+_10560147 | 0.09 |
ENST00000422133.1 |
HPCAL1 |
hippocalcin-like 1 |
| chr5_-_88119580 | 0.09 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr19_-_6670128 | 0.09 |
ENST00000245912.3 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
| chr1_+_161551101 | 0.09 |
ENST00000367962.4 ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B |
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr19_-_20748541 | 0.09 |
ENST00000427401.4 ENST00000594419.1 |
ZNF737 |
zinc finger protein 737 |
| chr10_+_35484053 | 0.09 |
ENST00000487763.1 ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM |
cAMP responsive element modulator |
| chr21_-_45078019 | 0.09 |
ENST00000542962.1 |
HSF2BP |
heat shock transcription factor 2 binding protein |
| chr22_-_50312052 | 0.09 |
ENST00000330817.6 |
ALG12 |
ALG12, alpha-1,6-mannosyltransferase |
| chr6_+_83073334 | 0.09 |
ENST00000369750.3 |
TPBG |
trophoblast glycoprotein |
| chr11_-_10715287 | 0.09 |
ENST00000423302.2 |
MRVI1 |
murine retrovirus integration site 1 homolog |
| chr20_+_4702548 | 0.09 |
ENST00000305817.2 |
PRND |
prion protein 2 (dublet) |
| chr15_+_67430339 | 0.09 |
ENST00000439724.3 |
SMAD3 |
SMAD family member 3 |
| chr19_+_41856816 | 0.08 |
ENST00000539627.1 |
TMEM91 |
transmembrane protein 91 |
| chr16_-_21877629 | 0.08 |
ENST00000544693.1 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chr6_+_126240442 | 0.08 |
ENST00000448104.1 ENST00000438495.1 ENST00000444128.1 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr1_-_241520385 | 0.08 |
ENST00000366564.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr12_+_133758115 | 0.08 |
ENST00000541009.2 ENST00000592241.1 |
ZNF268 |
zinc finger protein 268 |
| chr1_+_152627927 | 0.08 |
ENST00000444515.1 ENST00000536536.1 |
LINC00302 |
long intergenic non-protein coding RNA 302 |
| chr12_-_12503156 | 0.08 |
ENST00000543314.1 ENST00000396349.3 |
MANSC1 |
MANSC domain containing 1 |
| chr6_-_131277510 | 0.08 |
ENST00000525193.1 ENST00000527659.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr11_-_10715502 | 0.08 |
ENST00000547195.1 |
MRVI1 |
murine retrovirus integration site 1 homolog |
| chr16_+_28943260 | 0.08 |
ENST00000538922.1 ENST00000324662.3 ENST00000567541.1 |
CD19 |
CD19 molecule |
| chr2_-_202562716 | 0.08 |
ENST00000428900.2 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr8_+_42396712 | 0.08 |
ENST00000518574.1 ENST00000417410.2 ENST00000414154.2 |
SMIM19 |
small integral membrane protein 19 |
| chr2_-_37899323 | 0.08 |
ENST00000295324.3 ENST00000457889.1 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
| chr15_-_78526855 | 0.08 |
ENST00000541759.1 ENST00000558130.1 |
ACSBG1 |
acyl-CoA synthetase bubblegum family member 1 |
| chr15_-_63448973 | 0.08 |
ENST00000462430.1 |
RPS27L |
ribosomal protein S27-like |
| chr12_-_71533055 | 0.08 |
ENST00000552128.1 |
TSPAN8 |
tetraspanin 8 |
| chr5_+_159436120 | 0.07 |
ENST00000522793.1 ENST00000231238.5 |
TTC1 |
tetratricopeptide repeat domain 1 |
| chr4_-_178363581 | 0.07 |
ENST00000264595.2 |
AGA |
aspartylglucosaminidase |
| chr2_-_27712583 | 0.07 |
ENST00000260570.3 ENST00000359466.6 ENST00000416524.2 |
IFT172 |
intraflagellar transport 172 homolog (Chlamydomonas) |
| chr17_-_42295870 | 0.07 |
ENST00000526094.1 ENST00000529383.1 ENST00000530828.1 |
UBTF |
upstream binding transcription factor, RNA polymerase I |
| chr2_-_85829780 | 0.07 |
ENST00000334462.5 |
TMEM150A |
transmembrane protein 150A |
| chr1_-_21606013 | 0.07 |
ENST00000357071.4 |
ECE1 |
endothelin converting enzyme 1 |
| chr2_-_85829811 | 0.07 |
ENST00000306353.3 |
TMEM150A |
transmembrane protein 150A |
| chr7_-_142207004 | 0.07 |
ENST00000426318.2 |
TRBV10-2 |
T cell receptor beta variable 10-2 |
| chr5_-_61031495 | 0.07 |
ENST00000506550.1 ENST00000512882.2 |
CTD-2170G1.2 |
CTD-2170G1.2 |
| chr10_-_65225641 | 0.07 |
ENST00000399262.2 |
JMJD1C |
jumonji domain containing 1C |
| chr3_-_178984759 | 0.07 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr18_+_9136758 | 0.07 |
ENST00000383440.2 ENST00000262126.4 ENST00000577992.1 |
ANKRD12 |
ankyrin repeat domain 12 |
| chrX_+_78200913 | 0.07 |
ENST00000171757.2 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
| chr2_+_179316163 | 0.07 |
ENST00000409117.3 |
DFNB59 |
deafness, autosomal recessive 59 |
| chr5_+_118788433 | 0.07 |
ENST00000414835.2 |
HSD17B4 |
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr20_-_39928756 | 0.07 |
ENST00000432768.2 |
ZHX3 |
zinc fingers and homeoboxes 3 |
| chr14_-_34420259 | 0.07 |
ENST00000250457.3 ENST00000547327.2 |
EGLN3 |
egl-9 family hypoxia-inducible factor 3 |
| chr17_-_37844267 | 0.07 |
ENST00000579146.1 ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3 |
post-GPI attachment to proteins 3 |
| chrX_+_78200829 | 0.06 |
ENST00000544091.1 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
| chr12_+_12878829 | 0.06 |
ENST00000326765.6 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr17_-_29641084 | 0.06 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr14_+_88471468 | 0.06 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr9_-_134406565 | 0.06 |
ENST00000372210.3 ENST00000372211.3 |
UCK1 |
uridine-cytidine kinase 1 |
| chr14_-_96830207 | 0.06 |
ENST00000359933.4 |
ATG2B |
autophagy related 2B |
| chr8_+_136469684 | 0.06 |
ENST00000355849.5 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
| chr13_+_49280951 | 0.06 |
ENST00000282018.3 |
CYSLTR2 |
cysteinyl leukotriene receptor 2 |
| chr2_+_162016916 | 0.06 |
ENST00000405852.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr9_-_99616642 | 0.06 |
ENST00000478850.1 ENST00000481138.1 |
ZNF782 |
zinc finger protein 782 |
| chr2_-_220025548 | 0.06 |
ENST00000356853.5 |
NHEJ1 |
nonhomologous end-joining factor 1 |
| chr2_-_3521518 | 0.06 |
ENST00000382093.5 |
ADI1 |
acireductone dioxygenase 1 |
| chr11_+_103907308 | 0.06 |
ENST00000302259.3 |
DDI1 |
DNA-damage inducible 1 homolog 1 (S. cerevisiae) |
| chr3_-_88108212 | 0.06 |
ENST00000482016.1 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr1_-_12946025 | 0.06 |
ENST00000235349.5 |
PRAMEF4 |
PRAME family member 4 |
| chr15_+_74466744 | 0.06 |
ENST00000560862.1 ENST00000395118.1 |
ISLR |
immunoglobulin superfamily containing leucine-rich repeat |
| chr2_+_37571845 | 0.06 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr22_+_44577724 | 0.06 |
ENST00000466375.2 |
PARVG |
parvin, gamma |
| chr1_+_148560843 | 0.06 |
ENST00000442702.2 ENST00000369187.3 |
NBPF15 |
neuroblastoma breakpoint family, member 15 |
| chr6_+_27925019 | 0.06 |
ENST00000244623.1 |
OR2B6 |
olfactory receptor, family 2, subfamily B, member 6 |
| chr5_-_42811986 | 0.06 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr8_+_42396936 | 0.06 |
ENST00000416469.2 |
SMIM19 |
small integral membrane protein 19 |
| chr9_-_134406611 | 0.06 |
ENST00000372208.3 ENST00000372215.4 |
UCK1 |
uridine-cytidine kinase 1 |
| chr21_+_40823753 | 0.06 |
ENST00000333634.4 |
SH3BGR |
SH3 domain binding glutamic acid-rich protein |
| chr5_-_94417339 | 0.06 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr10_-_32667660 | 0.06 |
ENST00000375110.2 |
EPC1 |
enhancer of polycomb homolog 1 (Drosophila) |
| chr2_-_85829496 | 0.06 |
ENST00000409668.1 |
TMEM150A |
transmembrane protein 150A |
| chr7_+_65552756 | 0.06 |
ENST00000450043.1 |
AC068533.7 |
AC068533.7 |
| chr1_-_47407111 | 0.05 |
ENST00000371904.4 |
CYP4A11 |
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr16_+_22516172 | 0.05 |
ENST00000543407.1 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr21_-_35884573 | 0.05 |
ENST00000399286.2 |
KCNE1 |
potassium voltage-gated channel, Isk-related family, member 1 |
| chr1_+_161136180 | 0.05 |
ENST00000352210.5 ENST00000367999.4 ENST00000544598.1 ENST00000535223.1 ENST00000432542.2 |
PPOX |
protoporphyrinogen oxidase |
| chr2_-_121223697 | 0.05 |
ENST00000593290.1 |
FLJ14816 |
long intergenic non-protein coding RNA 1101 |
| chr1_-_153283194 | 0.05 |
ENST00000290722.1 |
PGLYRP3 |
peptidoglycan recognition protein 3 |
| chr11_+_60223225 | 0.05 |
ENST00000524807.1 ENST00000345732.4 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr1_-_57431679 | 0.05 |
ENST00000371237.4 ENST00000535057.1 ENST00000543257.1 |
C8B |
complement component 8, beta polypeptide |
| chr14_-_24701539 | 0.05 |
ENST00000534348.1 ENST00000524927.1 ENST00000250495.5 |
NEDD8-MDP1 NEDD8 |
NEDD8-MDP1 readthrough neural precursor cell expressed, developmentally down-regulated 8 |
| chr11_+_22689648 | 0.05 |
ENST00000278187.3 |
GAS2 |
growth arrest-specific 2 |
| chr14_+_45431379 | 0.05 |
ENST00000361577.3 ENST00000361462.2 ENST00000382233.2 |
FAM179B |
family with sequence similarity 179, member B |
| chr15_+_42694573 | 0.05 |
ENST00000397200.4 ENST00000569827.1 |
CAPN3 |
calpain 3, (p94) |
| chr22_+_22749343 | 0.05 |
ENST00000390298.2 |
IGLV7-43 |
immunoglobulin lambda variable 7-43 |
| chrX_-_153979315 | 0.05 |
ENST00000369575.3 ENST00000369568.4 ENST00000424127.2 |
GAB3 |
GRB2-associated binding protein 3 |
| chr12_-_7261772 | 0.05 |
ENST00000545280.1 ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL |
complement component 1, r subcomponent-like |
| chr1_+_158901329 | 0.05 |
ENST00000368140.1 ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
| chr6_-_149969829 | 0.05 |
ENST00000367411.2 |
KATNA1 |
katanin p60 (ATPase containing) subunit A 1 |
| chr10_-_65225722 | 0.05 |
ENST00000399251.1 |
JMJD1C |
jumonji domain containing 1C |
| chr6_-_149969871 | 0.05 |
ENST00000335643.8 ENST00000444282.1 |
KATNA1 |
katanin p60 (ATPase containing) subunit A 1 |
| chr19_-_12444491 | 0.05 |
ENST00000293725.5 |
ZNF563 |
zinc finger protein 563 |
| chr1_-_57285038 | 0.05 |
ENST00000343433.6 |
C1orf168 |
chromosome 1 open reading frame 168 |
| chr19_-_37663332 | 0.05 |
ENST00000392157.2 |
ZNF585A |
zinc finger protein 585A |
| chr17_-_5138099 | 0.05 |
ENST00000571800.1 ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP |
SLP adaptor and CSK interacting membrane protein |
| chr15_+_74466012 | 0.05 |
ENST00000249842.3 |
ISLR |
immunoglobulin superfamily containing leucine-rich repeat |
| chr11_-_106889157 | 0.05 |
ENST00000282249.2 |
GUCY1A2 |
guanylate cyclase 1, soluble, alpha 2 |
| chr4_+_72204755 | 0.05 |
ENST00000512686.1 ENST00000340595.3 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chrX_+_148622138 | 0.05 |
ENST00000450602.2 ENST00000441248.1 |
CXorf40A |
chromosome X open reading frame 40A |
| chr3_-_169587621 | 0.05 |
ENST00000523069.1 ENST00000316428.5 ENST00000264676.5 |
LRRC31 |
leucine rich repeat containing 31 |
| chr1_+_222886694 | 0.05 |
ENST00000426638.1 ENST00000537020.1 ENST00000539697.1 |
BROX |
BRO1 domain and CAAX motif containing |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.2 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.1 | 0.7 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.3 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 0.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.2 | GO:0002605 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.1 | 0.2 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.5 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.5 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.3 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.3 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.0 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.0 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.0 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.0 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.2 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.0 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |