Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
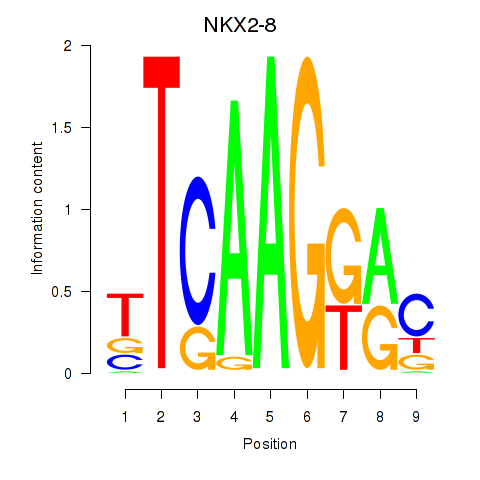
Results for NKX2-8
Z-value: 0.66
Transcription factors associated with NKX2-8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-8
|
ENSG00000136327.6 | NKX2-8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-8 | hg19_v2_chr14_-_37051798_37051831 | 0.11 | 7.9e-01 | Click! |
Activity profile of NKX2-8 motif
Sorted Z-values of NKX2-8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_92413353 | 0.62 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr5_+_92919043 | 0.56 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr16_+_88493879 | 0.43 |
ENST00000565624.1 ENST00000437464.1 |
ZNF469 |
zinc finger protein 469 |
| chr2_-_68547061 | 0.41 |
ENST00000263655.3 |
CNRIP1 |
cannabinoid receptor interacting protein 1 |
| chr16_-_3350614 | 0.30 |
ENST00000268674.2 |
TIGD7 |
tigger transposable element derived 7 |
| chr12_+_60058458 | 0.29 |
ENST00000548610.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr6_-_132272504 | 0.28 |
ENST00000367976.3 |
CTGF |
connective tissue growth factor |
| chr18_-_56296182 | 0.27 |
ENST00000361673.3 |
ALPK2 |
alpha-kinase 2 |
| chr3_-_58613323 | 0.24 |
ENST00000474531.1 ENST00000465970.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr5_-_95158644 | 0.23 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr8_+_27182862 | 0.23 |
ENST00000521164.1 ENST00000346049.5 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr19_-_51336443 | 0.22 |
ENST00000598673.1 |
KLK15 |
kallikrein-related peptidase 15 |
| chr3_-_8811288 | 0.22 |
ENST00000316793.3 ENST00000431493.1 |
OXTR |
oxytocin receptor |
| chr3_-_114790179 | 0.21 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr1_+_180165672 | 0.21 |
ENST00000443059.1 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
| chr3_-_114343768 | 0.18 |
ENST00000393785.2 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr4_-_119274121 | 0.18 |
ENST00000296498.3 |
PRSS12 |
protease, serine, 12 (neurotrypsin, motopsin) |
| chr14_-_107219365 | 0.17 |
ENST00000424969.2 |
IGHV3-74 |
immunoglobulin heavy variable 3-74 |
| chr19_-_18391708 | 0.16 |
ENST00000600972.1 |
JUND |
jun D proto-oncogene |
| chr6_-_87804815 | 0.15 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr6_-_137113604 | 0.14 |
ENST00000359015.4 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
| chr1_-_68698222 | 0.14 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr3_-_16524357 | 0.14 |
ENST00000432519.1 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr14_-_106586656 | 0.14 |
ENST00000390602.2 |
IGHV3-13 |
immunoglobulin heavy variable 3-13 |
| chr1_-_27998689 | 0.14 |
ENST00000339145.4 ENST00000362020.4 ENST00000361157.6 |
IFI6 |
interferon, alpha-inducible protein 6 |
| chr3_-_121740969 | 0.14 |
ENST00000393631.1 ENST00000273691.3 ENST00000344209.5 |
ILDR1 |
immunoglobulin-like domain containing receptor 1 |
| chr1_+_226013047 | 0.14 |
ENST00000366837.4 |
EPHX1 |
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr17_-_33390667 | 0.13 |
ENST00000378516.2 ENST00000268850.7 ENST00000394597.2 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr8_-_29120580 | 0.13 |
ENST00000524189.1 |
KIF13B |
kinesin family member 13B |
| chr4_+_37245799 | 0.13 |
ENST00000309447.5 |
KIAA1239 |
KIAA1239 |
| chr22_-_32341336 | 0.13 |
ENST00000248984.3 |
C22orf24 |
chromosome 22 open reading frame 24 |
| chr6_+_41010293 | 0.13 |
ENST00000373161.1 ENST00000373158.2 ENST00000470917.1 |
TSPO2 |
translocator protein 2 |
| chr19_-_9003586 | 0.12 |
ENST00000380951.5 |
MUC16 |
mucin 16, cell surface associated |
| chr18_-_44336998 | 0.12 |
ENST00000315087.7 |
ST8SIA5 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr7_-_137686791 | 0.12 |
ENST00000452463.1 ENST00000330387.6 ENST00000456390.1 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
| chr6_+_84222194 | 0.11 |
ENST00000536636.1 |
PRSS35 |
protease, serine, 35 |
| chr17_+_76227391 | 0.11 |
ENST00000586400.1 ENST00000421688.1 ENST00000374946.3 |
TMEM235 |
transmembrane protein 235 |
| chr19_-_38720354 | 0.11 |
ENST00000416611.1 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
| chr16_+_226658 | 0.11 |
ENST00000320868.5 ENST00000397797.1 |
HBA1 |
hemoglobin, alpha 1 |
| chr14_-_106518922 | 0.11 |
ENST00000390598.2 |
IGHV3-7 |
immunoglobulin heavy variable 3-7 |
| chr3_-_73673991 | 0.11 |
ENST00000308537.4 ENST00000263666.4 |
PDZRN3 |
PDZ domain containing ring finger 3 |
| chr20_-_44485835 | 0.10 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr15_-_56757329 | 0.10 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr6_+_84222220 | 0.10 |
ENST00000369700.3 |
PRSS35 |
protease, serine, 35 |
| chrX_-_151143140 | 0.10 |
ENST00000393914.3 ENST00000370328.3 ENST00000370325.1 |
GABRE |
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr2_-_106054952 | 0.10 |
ENST00000336660.5 ENST00000393352.3 ENST00000607522.1 |
FHL2 |
four and a half LIM domains 2 |
| chr14_-_106692191 | 0.10 |
ENST00000390607.2 |
IGHV3-21 |
immunoglobulin heavy variable 3-21 |
| chr3_-_114343039 | 0.10 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr13_-_23949671 | 0.09 |
ENST00000402364.1 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr1_+_228337553 | 0.09 |
ENST00000366714.2 |
GJC2 |
gap junction protein, gamma 2, 47kDa |
| chr10_+_115674530 | 0.09 |
ENST00000451472.1 |
AL162407.1 |
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr3_-_122134882 | 0.09 |
ENST00000330689.4 |
WDR5B |
WD repeat domain 5B |
| chr19_-_11347173 | 0.09 |
ENST00000587656.1 |
DOCK6 |
dedicator of cytokinesis 6 |
| chrX_-_64196351 | 0.09 |
ENST00000374839.3 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr12_-_118406777 | 0.09 |
ENST00000339824.5 |
KSR2 |
kinase suppressor of ras 2 |
| chr14_+_58894141 | 0.08 |
ENST00000423743.3 |
KIAA0586 |
KIAA0586 |
| chr2_+_62132800 | 0.08 |
ENST00000538736.1 |
COMMD1 |
copper metabolism (Murr1) domain containing 1 |
| chr12_-_75603236 | 0.08 |
ENST00000540018.1 |
KCNC2 |
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr2_-_152118352 | 0.08 |
ENST00000331426.5 |
RBM43 |
RNA binding motif protein 43 |
| chr14_+_58894103 | 0.08 |
ENST00000354386.6 ENST00000556134.1 |
KIAA0586 |
KIAA0586 |
| chr17_-_8198636 | 0.08 |
ENST00000577745.1 ENST00000579192.1 ENST00000396278.1 |
SLC25A35 |
solute carrier family 25, member 35 |
| chr3_-_178984759 | 0.08 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr17_+_76210367 | 0.08 |
ENST00000592734.1 ENST00000587746.1 |
BIRC5 |
baculoviral IAP repeat containing 5 |
| chr14_-_106994333 | 0.08 |
ENST00000390624.2 |
IGHV3-48 |
immunoglobulin heavy variable 3-48 |
| chr1_-_104238912 | 0.08 |
ENST00000330330.5 |
AMY1B |
amylase, alpha 1B (salivary) |
| chrX_-_64196307 | 0.08 |
ENST00000545618.1 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chrX_-_64196376 | 0.07 |
ENST00000447788.2 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr20_-_57089934 | 0.07 |
ENST00000439429.1 ENST00000371149.3 |
APCDD1L |
adenomatosis polyposis coli down-regulated 1-like |
| chr2_-_3504587 | 0.07 |
ENST00000415131.1 |
ADI1 |
acireductone dioxygenase 1 |
| chr3_+_187930719 | 0.07 |
ENST00000312675.4 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
| chr2_+_62132781 | 0.07 |
ENST00000311832.5 |
COMMD1 |
copper metabolism (Murr1) domain containing 1 |
| chr10_-_135171178 | 0.07 |
ENST00000368551.1 |
FUOM |
fucose mutarotase |
| chr15_-_75248954 | 0.07 |
ENST00000499788.2 |
RPP25 |
ribonuclease P/MRP 25kDa subunit |
| chr1_+_43291220 | 0.07 |
ENST00000372514.3 |
ERMAP |
erythroblast membrane-associated protein (Scianna blood group) |
| chr1_+_154401791 | 0.07 |
ENST00000476006.1 |
IL6R |
interleukin 6 receptor |
| chr12_-_102591604 | 0.07 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr16_-_67493110 | 0.07 |
ENST00000602876.1 |
ATP6V0D1 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chrX_+_48681768 | 0.07 |
ENST00000430858.1 |
HDAC6 |
histone deacetylase 6 |
| chr5_+_43602750 | 0.07 |
ENST00000505678.2 ENST00000512422.1 ENST00000264663.5 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr16_+_28875126 | 0.07 |
ENST00000359285.5 ENST00000538342.1 |
SH2B1 |
SH2B adaptor protein 1 |
| chr15_-_50558223 | 0.07 |
ENST00000267845.3 |
HDC |
histidine decarboxylase |
| chrX_-_69479654 | 0.07 |
ENST00000374519.2 |
P2RY4 |
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr17_+_58755184 | 0.07 |
ENST00000589222.1 ENST00000407086.3 ENST00000390652.5 |
BCAS3 |
breast carcinoma amplified sequence 3 |
| chr15_-_35047166 | 0.07 |
ENST00000290374.4 |
GJD2 |
gap junction protein, delta 2, 36kDa |
| chr12_-_75603482 | 0.07 |
ENST00000341669.3 ENST00000298972.1 ENST00000350228.2 |
KCNC2 |
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr22_-_37213554 | 0.07 |
ENST00000443735.1 |
PVALB |
parvalbumin |
| chr12_-_75603643 | 0.07 |
ENST00000549446.1 |
KCNC2 |
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr7_+_99816859 | 0.07 |
ENST00000317271.2 |
PVRIG |
poliovirus receptor related immunoglobulin domain containing |
| chr2_-_207024233 | 0.06 |
ENST00000423725.1 ENST00000233190.6 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr14_-_107131560 | 0.06 |
ENST00000390632.2 |
IGHV3-66 |
immunoglobulin heavy variable 3-66 |
| chr9_-_22009297 | 0.06 |
ENST00000276925.6 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr7_+_100318423 | 0.06 |
ENST00000252723.2 |
EPO |
erythropoietin |
| chr14_-_106573756 | 0.06 |
ENST00000390601.2 |
IGHV3-11 |
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr1_+_55464600 | 0.06 |
ENST00000371265.4 |
BSND |
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr7_-_92848878 | 0.06 |
ENST00000341723.4 |
HEPACAM2 |
HEPACAM family member 2 |
| chr7_-_92848858 | 0.06 |
ENST00000440868.1 |
HEPACAM2 |
HEPACAM family member 2 |
| chr16_-_18468926 | 0.06 |
ENST00000545114.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr16_+_8807419 | 0.06 |
ENST00000565016.1 ENST00000567812.1 |
ABAT |
4-aminobutyrate aminotransferase |
| chr13_+_108921977 | 0.06 |
ENST00000430559.1 ENST00000375887.4 |
TNFSF13B |
tumor necrosis factor (ligand) superfamily, member 13b |
| chr16_+_22501658 | 0.06 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chrX_-_48858667 | 0.06 |
ENST00000376423.4 ENST00000376441.1 |
GRIPAP1 |
GRIP1 associated protein 1 |
| chr19_-_51893827 | 0.05 |
ENST00000574814.1 |
CTD-2616J11.4 |
chromosome 19 open reading frame 84 |
| chr17_+_62503147 | 0.05 |
ENST00000553412.1 |
CEP95 |
centrosomal protein 95kDa |
| chr19_-_3600549 | 0.05 |
ENST00000589966.1 |
TBXA2R |
thromboxane A2 receptor |
| chr3_+_63953415 | 0.05 |
ENST00000484332.1 |
ATXN7 |
ataxin 7 |
| chr19_-_39322299 | 0.05 |
ENST00000601094.1 ENST00000595567.1 ENST00000602115.1 ENST00000601778.1 ENST00000597205.1 ENST00000595470.1 |
ECH1 |
enoyl CoA hydratase 1, peroxisomal |
| chr11_+_71791849 | 0.05 |
ENST00000423494.2 ENST00000539587.1 ENST00000538478.1 ENST00000324866.7 ENST00000439209.1 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
| chr15_+_51973680 | 0.05 |
ENST00000542355.2 |
SCG3 |
secretogranin III |
| chr11_+_71791693 | 0.05 |
ENST00000289488.2 ENST00000447974.1 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
| chr14_+_76618242 | 0.05 |
ENST00000557542.1 ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L |
G patch domain containing 2-like |
| chr16_+_32077386 | 0.05 |
ENST00000354689.6 |
IGHV3OR16-9 |
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr6_+_111408698 | 0.05 |
ENST00000368851.5 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr14_+_58894706 | 0.05 |
ENST00000261244.5 |
KIAA0586 |
KIAA0586 |
| chrX_-_53461288 | 0.05 |
ENST00000375298.4 ENST00000375304.5 |
HSD17B10 |
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr3_-_47950745 | 0.05 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr12_-_75603202 | 0.05 |
ENST00000393288.2 |
KCNC2 |
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr21_+_33671160 | 0.05 |
ENST00000303645.5 |
MRAP |
melanocortin 2 receptor accessory protein |
| chr8_-_145018905 | 0.05 |
ENST00000398774.2 |
PLEC |
plectin |
| chrX_-_15332665 | 0.04 |
ENST00000537676.1 ENST00000344384.4 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr17_+_76210267 | 0.04 |
ENST00000301633.4 ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5 |
baculoviral IAP repeat containing 5 |
| chr7_+_30811004 | 0.04 |
ENST00000265299.6 |
FAM188B |
family with sequence similarity 188, member B |
| chr9_+_35909478 | 0.04 |
ENST00000443779.1 |
LINC00961 |
long intergenic non-protein coding RNA 961 |
| chr9_-_21202204 | 0.04 |
ENST00000239347.3 |
IFNA7 |
interferon, alpha 7 |
| chr17_+_71228793 | 0.04 |
ENST00000426147.2 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chr14_-_107049312 | 0.04 |
ENST00000390627.2 |
IGHV3-53 |
immunoglobulin heavy variable 3-53 |
| chr3_+_137728842 | 0.04 |
ENST00000183605.5 |
CLDN18 |
claudin 18 |
| chr12_+_13349650 | 0.04 |
ENST00000256951.5 ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1 |
epithelial membrane protein 1 |
| chr7_-_92855762 | 0.04 |
ENST00000453812.2 ENST00000394468.2 |
HEPACAM2 |
HEPACAM family member 2 |
| chr9_+_96051469 | 0.04 |
ENST00000453718.1 |
WNK2 |
WNK lysine deficient protein kinase 2 |
| chr6_-_133084580 | 0.04 |
ENST00000525270.1 ENST00000530536.1 ENST00000524919.1 |
VNN2 |
vanin 2 |
| chr16_+_58426296 | 0.04 |
ENST00000426538.2 ENST00000328514.7 ENST00000318129.5 |
GINS3 |
GINS complex subunit 3 (Psf3 homolog) |
| chr14_-_106791536 | 0.04 |
ENST00000390613.2 |
IGHV3-30 |
immunoglobulin heavy variable 3-30 |
| chr10_+_118305435 | 0.04 |
ENST00000369221.2 |
PNLIP |
pancreatic lipase |
| chr7_-_127255724 | 0.04 |
ENST00000463946.1 ENST00000341640.2 |
PAX4 |
paired box 4 |
| chr15_+_51973550 | 0.04 |
ENST00000220478.3 |
SCG3 |
secretogranin III |
| chr14_-_106552755 | 0.04 |
ENST00000390600.2 |
IGHV3-9 |
immunoglobulin heavy variable 3-9 |
| chr1_+_41204506 | 0.03 |
ENST00000525290.1 ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC |
nuclear transcription factor Y, gamma |
| chrX_-_48776292 | 0.03 |
ENST00000376509.4 |
PIM2 |
pim-2 oncogene |
| chr7_+_30634297 | 0.03 |
ENST00000389266.3 |
GARS |
glycyl-tRNA synthetase |
| chr16_-_33647696 | 0.03 |
ENST00000558425.1 ENST00000569103.2 |
RP11-812E19.9 |
Uncharacterized protein |
| chr6_+_43968306 | 0.03 |
ENST00000442114.2 ENST00000336600.5 ENST00000439969.2 |
C6orf223 |
chromosome 6 open reading frame 223 |
| chr15_-_77363441 | 0.03 |
ENST00000346495.2 ENST00000424443.3 ENST00000561277.1 |
TSPAN3 |
tetraspanin 3 |
| chr7_+_73507409 | 0.03 |
ENST00000538333.3 |
LIMK1 |
LIM domain kinase 1 |
| chr20_-_43936937 | 0.03 |
ENST00000342716.4 ENST00000353917.5 ENST00000360607.6 ENST00000372751.4 |
MATN4 |
matrilin 4 |
| chr18_-_24722995 | 0.03 |
ENST00000581714.1 |
CHST9 |
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr16_-_66952779 | 0.03 |
ENST00000570262.1 ENST00000394055.3 ENST00000299752.4 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr14_+_58765103 | 0.03 |
ENST00000355431.3 ENST00000348476.3 ENST00000395168.3 |
ARID4A |
AT rich interactive domain 4A (RBP1-like) |
| chr8_+_23430157 | 0.03 |
ENST00000399967.3 |
FP15737 |
FP15737 |
| chr2_+_101591314 | 0.03 |
ENST00000450763.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr14_+_21498666 | 0.03 |
ENST00000481535.1 |
TPPP2 |
tubulin polymerization-promoting protein family member 2 |
| chr18_+_43304092 | 0.03 |
ENST00000321925.4 ENST00000587601.1 |
SLC14A1 |
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr17_-_46178527 | 0.03 |
ENST00000393408.3 |
CBX1 |
chromobox homolog 1 |
| chr12_+_16109519 | 0.03 |
ENST00000526530.1 |
DERA |
deoxyribose-phosphate aldolase (putative) |
| chr12_-_95397442 | 0.03 |
ENST00000547157.1 ENST00000547986.1 ENST00000327772.2 |
NDUFA12 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr16_+_88872176 | 0.03 |
ENST00000569140.1 |
CDT1 |
chromatin licensing and DNA replication factor 1 |
| chr2_+_119699742 | 0.03 |
ENST00000327097.4 |
MARCO |
macrophage receptor with collagenous structure |
| chr10_-_118032979 | 0.03 |
ENST00000355422.6 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr16_-_66952742 | 0.03 |
ENST00000565235.2 ENST00000568632.1 ENST00000565796.1 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr3_+_108855558 | 0.03 |
ENST00000467240.1 ENST00000477643.1 ENST00000479039.1 ENST00000593799.1 |
RP11-59E19.1 |
RP11-59E19.1 |
| chr19_-_39881669 | 0.03 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr6_+_34433844 | 0.02 |
ENST00000244458.2 ENST00000374043.2 |
PACSIN1 |
protein kinase C and casein kinase substrate in neurons 1 |
| chr15_-_77363375 | 0.02 |
ENST00000559494.1 |
TSPAN3 |
tetraspanin 3 |
| chr22_+_21213771 | 0.02 |
ENST00000439214.1 |
SNAP29 |
synaptosomal-associated protein, 29kDa |
| chr7_-_99277610 | 0.02 |
ENST00000343703.5 ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5 |
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr13_-_103346854 | 0.02 |
ENST00000267273.6 |
METTL21C |
methyltransferase like 21C |
| chr5_+_34929677 | 0.02 |
ENST00000342382.4 ENST00000382021.2 ENST00000303525.7 |
DNAJC21 |
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr17_-_9808887 | 0.02 |
ENST00000226193.5 |
RCVRN |
recoverin |
| chr12_+_121837905 | 0.02 |
ENST00000392465.3 ENST00000554606.1 ENST00000392464.2 ENST00000555076.1 |
RNF34 |
ring finger protein 34, E3 ubiquitin protein ligase |
| chr1_+_151512775 | 0.02 |
ENST00000368849.3 ENST00000392712.3 ENST00000353024.3 ENST00000368848.2 ENST00000538902.1 |
TUFT1 |
tuftelin 1 |
| chr6_+_69942298 | 0.02 |
ENST00000238918.8 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
| chr2_-_110371720 | 0.02 |
ENST00000356688.4 |
SEPT10 |
septin 10 |
| chr14_+_22309368 | 0.02 |
ENST00000390433.1 |
TRAV12-1 |
T cell receptor alpha variable 12-1 |
| chr19_-_39322497 | 0.02 |
ENST00000221418.4 |
ECH1 |
enoyl CoA hydratase 1, peroxisomal |
| chr20_-_48747662 | 0.02 |
ENST00000371656.2 |
TMEM189 |
transmembrane protein 189 |
| chr3_-_187455680 | 0.02 |
ENST00000438077.1 |
BCL6 |
B-cell CLL/lymphoma 6 |
| chr11_-_10828892 | 0.02 |
ENST00000525681.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr1_+_109289279 | 0.02 |
ENST00000370008.3 |
STXBP3 |
syntaxin binding protein 3 |
| chr6_+_32146131 | 0.02 |
ENST00000375094.3 |
RNF5 |
ring finger protein 5, E3 ubiquitin protein ligase |
| chr15_-_28419569 | 0.02 |
ENST00000569772.1 |
HERC2 |
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr21_-_35899113 | 0.02 |
ENST00000492600.1 ENST00000481448.1 ENST00000381132.2 |
RCAN1 |
regulator of calcineurin 1 |
| chr11_-_5364809 | 0.02 |
ENST00000300773.2 |
OR51B5 |
olfactory receptor, family 51, subfamily B, member 5 |
| chr16_+_33020496 | 0.02 |
ENST00000565407.2 |
IGHV3OR16-8 |
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr17_+_39994032 | 0.02 |
ENST00000293303.4 ENST00000438813.1 |
KLHL10 |
kelch-like family member 10 |
| chr1_-_115292591 | 0.02 |
ENST00000438362.2 |
CSDE1 |
cold shock domain containing E1, RNA-binding |
| chr3_+_39093481 | 0.01 |
ENST00000302313.5 ENST00000544962.1 ENST00000396258.3 ENST00000418020.1 |
WDR48 |
WD repeat domain 48 |
| chr18_-_72920372 | 0.01 |
ENST00000581620.1 ENST00000582437.1 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
| chr12_+_25205568 | 0.01 |
ENST00000548766.1 ENST00000556887.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr15_-_20170354 | 0.01 |
ENST00000338912.5 |
IGHV1OR15-9 |
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr19_+_51630287 | 0.01 |
ENST00000599948.1 |
SIGLEC9 |
sialic acid binding Ig-like lectin 9 |
| chr4_+_90816033 | 0.01 |
ENST00000264790.2 ENST00000394981.1 |
MMRN1 |
multimerin 1 |
| chr20_-_48530230 | 0.01 |
ENST00000422556.1 |
SPATA2 |
spermatogenesis associated 2 |
| chr15_+_97326659 | 0.01 |
ENST00000558553.1 |
SPATA8 |
spermatogenesis associated 8 |
| chr2_+_119699864 | 0.01 |
ENST00000541757.1 ENST00000412481.1 |
MARCO |
macrophage receptor with collagenous structure |
| chr5_+_140474181 | 0.01 |
ENST00000194155.4 |
PCDHB2 |
protocadherin beta 2 |
| chr14_-_107199464 | 0.01 |
ENST00000433072.2 |
IGHV3-72 |
immunoglobulin heavy variable 3-72 |
| chr2_-_136288740 | 0.01 |
ENST00000264159.6 ENST00000536680.1 |
ZRANB3 |
zinc finger, RAN-binding domain containing 3 |
| chr3_-_108836977 | 0.01 |
ENST00000232603.5 |
MORC1 |
MORC family CW-type zinc finger 1 |
| chr19_-_44259136 | 0.01 |
ENST00000270066.6 |
SMG9 |
SMG9 nonsense mediated mRNA decay factor |
| chr11_+_18720316 | 0.01 |
ENST00000280734.2 |
TMEM86A |
transmembrane protein 86A |
| chr14_+_37667193 | 0.01 |
ENST00000539062.2 |
MIPOL1 |
mirror-image polydactyly 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.2 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.3 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.3 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.1 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:0031296 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.0 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.0 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |