Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
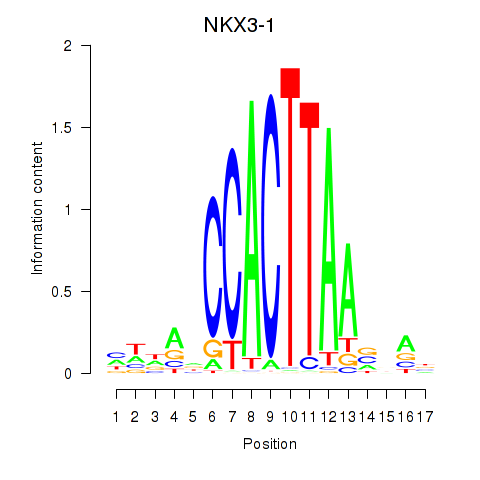
Results for NKX3-1
Z-value: 0.05
Transcription factors associated with NKX3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-1
|
ENSG00000167034.9 | NKX3-1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-1 | hg19_v2_chr8_-_23540402_23540466 | 0.78 | 2.3e-02 | Click! |
Activity profile of NKX3-1 motif
Sorted Z-values of NKX3-1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX3-1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_32264040 | 0.19 |
ENST00000398664.3 |
TP53TG3D |
TP53 target 3D |
| chr16_-_32688053 | 0.19 |
ENST00000398682.4 |
TP53TG3 |
TP53 target 3 |
| chr3_+_98482175 | 0.16 |
ENST00000485391.1 ENST00000492254.1 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr5_-_43412418 | 0.15 |
ENST00000537013.1 ENST00000361115.4 |
CCL28 |
chemokine (C-C motif) ligand 28 |
| chr4_-_76928641 | 0.14 |
ENST00000264888.5 |
CXCL9 |
chemokine (C-X-C motif) ligand 9 |
| chrX_+_85403487 | 0.12 |
ENST00000373125.4 |
DACH2 |
dachshund homolog 2 (Drosophila) |
| chr1_+_171217622 | 0.12 |
ENST00000433267.1 ENST00000367750.3 |
FMO1 |
flavin containing monooxygenase 1 |
| chrX_+_85403445 | 0.12 |
ENST00000373131.1 |
DACH2 |
dachshund homolog 2 (Drosophila) |
| chr9_-_73477826 | 0.11 |
ENST00000396285.1 ENST00000396292.4 ENST00000396280.5 ENST00000358082.3 ENST00000408909.2 ENST00000377097.3 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr16_+_33204156 | 0.10 |
ENST00000398667.4 |
TP53TG3C |
TP53 target 3C |
| chr14_+_95078714 | 0.10 |
ENST00000393078.3 ENST00000393080.4 ENST00000467132.1 |
SERPINA3 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr6_+_80129989 | 0.10 |
ENST00000429444.1 |
RP1-232L24.3 |
RP1-232L24.3 |
| chr4_-_170924888 | 0.09 |
ENST00000502832.1 ENST00000393704.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr14_-_60337684 | 0.09 |
ENST00000267484.5 |
RTN1 |
reticulon 1 |
| chr20_-_56285595 | 0.08 |
ENST00000395816.3 ENST00000347215.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr11_+_7506713 | 0.08 |
ENST00000329293.3 ENST00000534244.1 |
OLFML1 |
olfactomedin-like 1 |
| chr5_+_150404904 | 0.08 |
ENST00000521632.1 |
GPX3 |
glutathione peroxidase 3 (plasma) |
| chr20_-_56286479 | 0.08 |
ENST00000265626.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr19_+_50433476 | 0.07 |
ENST00000596658.1 |
ATF5 |
activating transcription factor 5 |
| chr3_-_18480260 | 0.07 |
ENST00000454909.2 |
SATB1 |
SATB homeobox 1 |
| chr3_-_178976996 | 0.07 |
ENST00000485523.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr6_+_26124373 | 0.07 |
ENST00000377791.2 ENST00000602637.1 |
HIST1H2AC |
histone cluster 1, H2ac |
| chr17_-_34257731 | 0.06 |
ENST00000431884.2 ENST00000425909.3 ENST00000394528.3 ENST00000430160.2 |
RDM1 |
RAD52 motif 1 |
| chr19_-_42894420 | 0.06 |
ENST00000597255.1 ENST00000222032.5 |
CNFN |
cornifelin |
| chr1_-_48866517 | 0.06 |
ENST00000371841.1 |
SPATA6 |
spermatogenesis associated 6 |
| chr17_-_2966901 | 0.06 |
ENST00000575751.1 |
OR1D5 |
olfactory receptor, family 1, subfamily D, member 5 |
| chr17_-_29641084 | 0.05 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr14_+_23654525 | 0.05 |
ENST00000399910.1 ENST00000492621.1 |
C14orf164 |
chromosome 14 open reading frame 164 |
| chr16_-_31076332 | 0.05 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chrX_-_153637612 | 0.05 |
ENST00000369807.1 ENST00000369808.3 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
| chr19_+_54609230 | 0.05 |
ENST00000420296.1 |
NDUFA3 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr17_-_76870126 | 0.05 |
ENST00000586057.1 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr5_-_139937895 | 0.05 |
ENST00000336283.6 |
SRA1 |
steroid receptor RNA activator 1 |
| chr16_-_87970122 | 0.05 |
ENST00000309893.2 |
CA5A |
carbonic anhydrase VA, mitochondrial |
| chr19_+_35396219 | 0.05 |
ENST00000595783.1 |
CTC-523E23.6 |
CTC-523E23.6 |
| chr17_-_76870222 | 0.05 |
ENST00000585421.1 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr16_-_31076273 | 0.05 |
ENST00000426488.2 |
ZNF668 |
zinc finger protein 668 |
| chr22_+_18632666 | 0.04 |
ENST00000215794.7 |
USP18 |
ubiquitin specific peptidase 18 |
| chr5_-_134783038 | 0.04 |
ENST00000503143.2 |
C5orf20 |
chromosome 5 open reading frame 20 |
| chr3_+_156807663 | 0.04 |
ENST00000467995.1 ENST00000474477.1 ENST00000471719.1 |
LINC00881 |
long intergenic non-protein coding RNA 881 |
| chr12_-_92536433 | 0.04 |
ENST00000551563.2 ENST00000546975.1 ENST00000549802.1 |
C12orf79 |
chromosome 12 open reading frame 79 |
| chr19_-_42006539 | 0.04 |
ENST00000597702.1 ENST00000588495.1 ENST00000595837.1 ENST00000594315.1 |
AC011526.1 |
AC011526.1 |
| chr17_-_55911970 | 0.04 |
ENST00000581805.1 ENST00000580960.1 |
RP11-60A24.3 |
RP11-60A24.3 |
| chr18_+_61144160 | 0.04 |
ENST00000489441.1 ENST00000424602.1 |
SERPINB5 |
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr11_+_9482512 | 0.04 |
ENST00000396602.2 ENST00000530463.1 ENST00000533542.1 ENST00000532577.1 ENST00000396597.3 |
ZNF143 |
zinc finger protein 143 |
| chr2_-_187367356 | 0.04 |
ENST00000595956.1 |
AC018867.2 |
AC018867.2 |
| chr4_-_138453606 | 0.04 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr7_-_112635675 | 0.04 |
ENST00000447785.1 ENST00000451962.1 |
AC018464.3 |
AC018464.3 |
| chr8_+_27168988 | 0.04 |
ENST00000397501.1 ENST00000338238.4 ENST00000544172.1 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr3_+_158991025 | 0.04 |
ENST00000337808.6 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr17_-_7145475 | 0.04 |
ENST00000571129.1 ENST00000571253.1 ENST00000573928.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr17_-_39526052 | 0.04 |
ENST00000251646.3 |
KRT33B |
keratin 33B |
| chr10_+_120116527 | 0.04 |
ENST00000445161.1 |
LINC00867 |
long intergenic non-protein coding RNA 867 |
| chr4_-_46126093 | 0.04 |
ENST00000295452.4 |
GABRG1 |
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr14_-_74551096 | 0.03 |
ENST00000350259.4 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr6_-_57087042 | 0.03 |
ENST00000317483.3 |
RAB23 |
RAB23, member RAS oncogene family |
| chr17_-_14683517 | 0.03 |
ENST00000379640.1 |
AC005863.1 |
AC005863.1 |
| chr15_+_58702742 | 0.03 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr10_-_91403625 | 0.03 |
ENST00000322191.6 ENST00000342512.3 ENST00000371774.2 |
PANK1 |
pantothenate kinase 1 |
| chr14_-_68066849 | 0.03 |
ENST00000558493.1 ENST00000561272.1 |
PIGH |
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr20_-_271009 | 0.03 |
ENST00000382369.5 |
C20orf96 |
chromosome 20 open reading frame 96 |
| chr10_+_5406935 | 0.03 |
ENST00000380433.3 |
UCN3 |
urocortin 3 |
| chr1_-_89488510 | 0.03 |
ENST00000564665.1 ENST00000370481.4 |
GBP3 |
guanylate binding protein 3 |
| chr1_-_160232312 | 0.03 |
ENST00000440682.1 |
DCAF8 |
DDB1 and CUL4 associated factor 8 |
| chr13_+_24144509 | 0.03 |
ENST00000248484.4 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr8_+_77593448 | 0.03 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr11_-_62323702 | 0.03 |
ENST00000530285.1 |
AHNAK |
AHNAK nucleoprotein |
| chr4_+_70916119 | 0.03 |
ENST00000246896.3 ENST00000511674.1 |
HTN1 |
histatin 1 |
| chrX_-_102757802 | 0.03 |
ENST00000372633.1 |
RAB40A |
RAB40A, member RAS oncogene family |
| chr3_+_132316081 | 0.03 |
ENST00000249887.2 |
ACKR4 |
atypical chemokine receptor 4 |
| chr15_+_31196080 | 0.03 |
ENST00000561607.1 ENST00000565466.1 |
FAN1 |
FANCD2/FANCI-associated nuclease 1 |
| chr5_+_140723601 | 0.03 |
ENST00000253812.6 |
PCDHGA3 |
protocadherin gamma subfamily A, 3 |
| chr13_+_50570019 | 0.03 |
ENST00000442421.1 |
TRIM13 |
tripartite motif containing 13 |
| chr19_+_58193388 | 0.03 |
ENST00000596085.1 ENST00000594684.1 |
ZNF551 AC003006.7 |
zinc finger protein 551 Uncharacterized protein |
| chr20_-_271304 | 0.03 |
ENST00000400269.3 ENST00000360321.2 |
C20orf96 |
chromosome 20 open reading frame 96 |
| chr15_+_44084040 | 0.03 |
ENST00000249786.4 |
SERF2 |
small EDRK-rich factor 2 |
| chr3_-_51813009 | 0.03 |
ENST00000398780.3 |
IQCF6 |
IQ motif containing F6 |
| chr4_-_48082192 | 0.03 |
ENST00000507351.1 |
TXK |
TXK tyrosine kinase |
| chr12_-_89920030 | 0.02 |
ENST00000413530.1 ENST00000547474.1 |
GALNT4 POC1B-GALNT4 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr6_+_63921399 | 0.02 |
ENST00000356170.3 |
FKBP1C |
FK506 binding protein 1C |
| chr5_+_112312416 | 0.02 |
ENST00000389063.2 |
DCP2 |
decapping mRNA 2 |
| chr4_-_1685718 | 0.02 |
ENST00000472884.2 ENST00000489363.1 ENST00000308132.6 ENST00000463238.1 |
FAM53A |
family with sequence similarity 53, member A |
| chr7_-_2883928 | 0.02 |
ENST00000275364.3 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
| chr20_-_45318051 | 0.02 |
ENST00000372102.3 |
TP53RK |
TP53 regulating kinase |
| chr20_-_2821271 | 0.02 |
ENST00000448755.1 ENST00000360652.2 |
PCED1A |
PC-esterase domain containing 1A |
| chr15_+_85147127 | 0.02 |
ENST00000541040.1 ENST00000538076.1 ENST00000485222.2 |
ZSCAN2 |
zinc finger and SCAN domain containing 2 |
| chr8_-_49833978 | 0.02 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chrX_-_107681633 | 0.02 |
ENST00000394872.2 ENST00000334504.7 |
COL4A6 |
collagen, type IV, alpha 6 |
| chr14_-_74551172 | 0.02 |
ENST00000553458.1 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chrX_+_153237740 | 0.02 |
ENST00000369982.4 |
TMEM187 |
transmembrane protein 187 |
| chr16_-_3074231 | 0.02 |
ENST00000572355.1 ENST00000248089.3 ENST00000574980.1 ENST00000354679.3 ENST00000396916.1 ENST00000573842.1 |
HCFC1R1 |
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr3_+_113251143 | 0.02 |
ENST00000264852.4 ENST00000393830.3 |
SIDT1 |
SID1 transmembrane family, member 1 |
| chr20_+_2795609 | 0.02 |
ENST00000554164.1 ENST00000380593.4 |
TMEM239 TMEM239 |
transmembrane protein 239 CDNA FLJ26142 fis, clone TST04526; Transmembrane protein 239; Uncharacterized protein |
| chr6_+_43266063 | 0.02 |
ENST00000372574.3 |
SLC22A7 |
solute carrier family 22 (organic anion transporter), member 7 |
| chr8_-_97247759 | 0.02 |
ENST00000518406.1 ENST00000523920.1 ENST00000287022.5 |
UQCRB |
ubiquinol-cytochrome c reductase binding protein |
| chr5_+_178977546 | 0.02 |
ENST00000319449.4 ENST00000377001.2 |
RUFY1 |
RUN and FYVE domain containing 1 |
| chr5_+_150051149 | 0.02 |
ENST00000523553.1 |
MYOZ3 |
myozenin 3 |
| chr5_-_150138551 | 0.02 |
ENST00000446090.2 ENST00000447998.2 |
DCTN4 |
dynactin 4 (p62) |
| chr8_-_49834299 | 0.02 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr20_-_2821756 | 0.02 |
ENST00000356872.3 ENST00000439542.1 |
PCED1A |
PC-esterase domain containing 1A |
| chr10_+_18629628 | 0.02 |
ENST00000377329.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr20_+_2821340 | 0.02 |
ENST00000380445.3 ENST00000380469.3 |
VPS16 |
vacuolar protein sorting 16 homolog (S. cerevisiae) |
| chr16_+_73420942 | 0.02 |
ENST00000554640.1 ENST00000562661.1 ENST00000561875.1 |
RP11-140I24.1 |
RP11-140I24.1 |
| chr17_-_7307358 | 0.02 |
ENST00000576017.1 ENST00000302422.3 ENST00000535512.1 |
TMEM256 TMEM256-PLSCR3 |
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr8_-_93029520 | 0.02 |
ENST00000521553.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr15_-_55563072 | 0.02 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr5_-_176889381 | 0.02 |
ENST00000393563.4 ENST00000512501.1 |
DBN1 |
drebrin 1 |
| chr15_-_43876702 | 0.02 |
ENST00000348806.6 |
PPIP5K1 |
diphosphoinositol pentakisphosphate kinase 1 |
| chr14_+_22580233 | 0.02 |
ENST00000390454.2 |
TRAV25 |
T cell receptor alpha variable 25 |
| chr12_+_111051902 | 0.02 |
ENST00000397655.3 ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1 |
tectonic family member 1 |
| chr11_+_108093839 | 0.02 |
ENST00000452508.2 |
ATM |
ataxia telangiectasia mutated |
| chr7_-_5998714 | 0.02 |
ENST00000539903.1 |
RSPH10B |
radial spoke head 10 homolog B (Chlamydomonas) |
| chr1_+_84873913 | 0.02 |
ENST00000370662.3 |
DNASE2B |
deoxyribonuclease II beta |
| chr2_+_201994569 | 0.02 |
ENST00000457277.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr1_+_27114589 | 0.02 |
ENST00000431541.1 ENST00000449950.2 ENST00000374145.1 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr2_+_113672770 | 0.02 |
ENST00000311328.2 |
IL37 |
interleukin 37 |
| chr1_-_120190396 | 0.02 |
ENST00000421812.2 |
ZNF697 |
zinc finger protein 697 |
| chr3_-_48632593 | 0.02 |
ENST00000454817.1 ENST00000328333.8 |
COL7A1 |
collagen, type VII, alpha 1 |
| chr15_+_44084503 | 0.02 |
ENST00000409960.2 ENST00000409646.1 ENST00000594896.1 ENST00000339624.5 ENST00000409291.1 ENST00000402131.1 ENST00000403425.1 ENST00000430901.1 |
SERF2 |
small EDRK-rich factor 2 |
| chr20_-_36661826 | 0.02 |
ENST00000373448.2 ENST00000373447.3 |
TTI1 |
TELO2 interacting protein 1 |
| chr10_-_61900762 | 0.02 |
ENST00000355288.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr12_+_96883347 | 0.02 |
ENST00000524981.4 ENST00000298953.3 |
C12orf55 |
chromosome 12 open reading frame 55 |
| chr17_+_2699697 | 0.02 |
ENST00000254695.8 ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
| chr12_-_89919765 | 0.02 |
ENST00000541909.1 ENST00000313546.3 |
POC1B |
POC1 centriolar protein B |
| chr13_+_24883703 | 0.02 |
ENST00000332018.4 |
C1QTNF9 |
C1q and tumor necrosis factor related protein 9 |
| chr11_+_67798090 | 0.02 |
ENST00000313468.5 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr13_+_115000556 | 0.02 |
ENST00000252458.6 |
CDC16 |
cell division cycle 16 |
| chr4_-_138453559 | 0.01 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr12_-_75603482 | 0.01 |
ENST00000341669.3 ENST00000298972.1 ENST00000350228.2 |
KCNC2 |
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr12_+_111051832 | 0.01 |
ENST00000550703.2 ENST00000551590.1 |
TCTN1 |
tectonic family member 1 |
| chr2_-_27545921 | 0.01 |
ENST00000402310.1 ENST00000405983.1 ENST00000403262.2 ENST00000428910.1 ENST00000402722.1 ENST00000399052.4 ENST00000380044.1 ENST00000405076.1 |
MPV17 |
MpV17 mitochondrial inner membrane protein |
| chr2_-_79313973 | 0.01 |
ENST00000454188.1 |
REG1B |
regenerating islet-derived 1 beta |
| chr19_+_58144529 | 0.01 |
ENST00000347302.3 ENST00000254182.7 ENST00000391703.3 ENST00000541801.1 ENST00000299871.5 ENST00000544273.1 |
ZNF211 |
zinc finger protein 211 |
| chr19_+_58144589 | 0.01 |
ENST00000240731.4 |
ZNF211 |
zinc finger protein 211 |
| chr19_+_52074502 | 0.01 |
ENST00000545217.1 ENST00000262259.2 ENST00000596504.1 |
ZNF175 |
zinc finger protein 175 |
| chr16_+_1832902 | 0.01 |
ENST00000262302.9 ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2 |
nucleotide binding protein 2 |
| chr5_-_1799932 | 0.01 |
ENST00000382647.7 ENST00000505059.2 |
MRPL36 |
mitochondrial ribosomal protein L36 |
| chr11_+_65266507 | 0.01 |
ENST00000544868.1 |
MALAT1 |
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr8_+_87878640 | 0.01 |
ENST00000518476.1 |
CNBD1 |
cyclic nucleotide binding domain containing 1 |
| chr19_-_2944907 | 0.01 |
ENST00000314531.4 |
ZNF77 |
zinc finger protein 77 |
| chr19_-_56110859 | 0.01 |
ENST00000221665.3 ENST00000592585.1 |
FIZ1 |
FLT3-interacting zinc finger 1 |
| chr11_+_8040739 | 0.01 |
ENST00000534099.1 |
TUB |
tubby bipartite transcription factor |
| chr11_-_61687739 | 0.01 |
ENST00000531922.1 ENST00000301773.5 |
RAB3IL1 |
RAB3A interacting protein (rabin3)-like 1 |
| chr1_+_161475208 | 0.01 |
ENST00000367972.4 ENST00000271450.6 |
FCGR2A |
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| chr17_+_37824411 | 0.01 |
ENST00000269582.2 |
PNMT |
phenylethanolamine N-methyltransferase |
| chr6_-_80247105 | 0.01 |
ENST00000369846.4 |
LCA5 |
Leber congenital amaurosis 5 |
| chr13_+_115000521 | 0.01 |
ENST00000252457.5 ENST00000375308.1 |
CDC16 |
cell division cycle 16 |
| chr11_+_67798114 | 0.01 |
ENST00000453471.2 ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr11_+_71791693 | 0.01 |
ENST00000289488.2 ENST00000447974.1 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
| chr3_-_160117301 | 0.01 |
ENST00000326448.7 ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80 |
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr4_+_155702496 | 0.01 |
ENST00000510397.1 |
RBM46 |
RNA binding motif protein 46 |
| chr5_-_76788134 | 0.01 |
ENST00000507029.1 |
WDR41 |
WD repeat domain 41 |
| chr10_+_114133773 | 0.01 |
ENST00000354655.4 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
| chr1_+_202431859 | 0.01 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr19_+_58193337 | 0.01 |
ENST00000601064.1 ENST00000282296.5 ENST00000356715.4 |
ZNF551 |
zinc finger protein 551 |
| chr17_+_37824217 | 0.01 |
ENST00000394246.1 |
PNMT |
phenylethanolamine N-methyltransferase |
| chr5_-_1799864 | 0.01 |
ENST00000510999.1 |
MRPL36 |
mitochondrial ribosomal protein L36 |
| chr6_-_144416737 | 0.01 |
ENST00000367569.2 |
SF3B5 |
splicing factor 3b, subunit 5, 10kDa |
| chr1_-_22109484 | 0.01 |
ENST00000529637.1 |
USP48 |
ubiquitin specific peptidase 48 |
| chr10_+_46994087 | 0.01 |
ENST00000374317.1 |
GPRIN2 |
G protein regulated inducer of neurite outgrowth 2 |
| chr5_+_125758865 | 0.01 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr1_-_154832316 | 0.01 |
ENST00000361147.4 |
KCNN3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr5_-_76788024 | 0.01 |
ENST00000515253.1 ENST00000414719.2 ENST00000507654.1 ENST00000514559.1 ENST00000511791.1 |
WDR41 |
WD repeat domain 41 |
| chr16_+_89628778 | 0.01 |
ENST00000472354.1 |
RPL13 |
ribosomal protein L13 |
| chr5_-_1799965 | 0.01 |
ENST00000508987.1 |
MRPL36 |
mitochondrial ribosomal protein L36 |
| chr5_+_125758813 | 0.01 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr15_-_43513187 | 0.01 |
ENST00000540029.1 ENST00000441366.2 |
EPB42 |
erythrocyte membrane protein band 4.2 |
| chr1_-_39339777 | 0.01 |
ENST00000397572.2 |
MYCBP |
MYC binding protein |
| chr20_+_3052264 | 0.01 |
ENST00000217386.2 |
OXT |
oxytocin/neurophysin I prepropeptide |
| chr11_+_8704748 | 0.01 |
ENST00000526562.1 ENST00000525981.1 |
RPL27A |
ribosomal protein L27a |
| chr3_+_100428188 | 0.01 |
ENST00000418917.2 ENST00000490574.1 |
TFG |
TRK-fused gene |
| chr11_-_71823796 | 0.01 |
ENST00000545680.1 ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr3_+_100428268 | 0.01 |
ENST00000240851.4 |
TFG |
TRK-fused gene |
| chr15_-_31393910 | 0.01 |
ENST00000397795.2 ENST00000256552.6 ENST00000559179.1 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
| chr4_-_105416039 | 0.01 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr8_-_30706608 | 0.01 |
ENST00000256246.2 |
TEX15 |
testis expressed 15 |
| chr1_-_35497506 | 0.01 |
ENST00000317538.5 ENST00000373340.2 ENST00000357182.4 |
ZMYM6 |
zinc finger, MYM-type 6 |
| chr4_+_76481258 | 0.01 |
ENST00000311623.4 ENST00000435974.2 |
C4orf26 |
chromosome 4 open reading frame 26 |
| chr17_+_80674559 | 0.01 |
ENST00000269373.6 ENST00000535965.1 ENST00000577128.1 ENST00000573158.1 |
FN3KRP |
fructosamine 3 kinase related protein |
| chr11_-_101454234 | 0.01 |
ENST00000348423.4 ENST00000360497.4 ENST00000532133.1 |
TRPC6 |
transient receptor potential cation channel, subfamily C, member 6 |
| chr20_-_49253425 | 0.01 |
ENST00000045083.2 |
FAM65C |
family with sequence similarity 65, member C |
| chr3_+_100428316 | 0.01 |
ENST00000479672.1 ENST00000476228.1 ENST00000463568.1 |
TFG |
TRK-fused gene |
| chr10_-_28571015 | 0.01 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr4_+_37962018 | 0.01 |
ENST00000504686.1 |
PTTG2 |
pituitary tumor-transforming 2 |
| chr6_+_43603552 | 0.01 |
ENST00000372171.4 |
MAD2L1BP |
MAD2L1 binding protein |
| chr7_+_150147711 | 0.01 |
ENST00000307271.3 |
GIMAP8 |
GTPase, IMAP family member 8 |
| chr17_-_64187973 | 0.01 |
ENST00000583358.1 ENST00000392769.2 |
CEP112 |
centrosomal protein 112kDa |
| chr10_+_135204338 | 0.01 |
ENST00000468317.2 |
RP11-108K14.8 |
Mitochondrial GTPase 1 |
| chr10_+_103348031 | 0.01 |
ENST00000370151.4 ENST00000370147.1 ENST00000370148.2 |
DPCD |
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr17_+_33914460 | 0.01 |
ENST00000537622.2 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr11_-_118901559 | 0.01 |
ENST00000330775.7 ENST00000545985.1 ENST00000357590.5 ENST00000538950.1 |
SLC37A4 |
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr3_-_69101461 | 0.01 |
ENST00000543976.1 |
TMF1 |
TATA element modulatory factor 1 |
| chr16_+_83986827 | 0.01 |
ENST00000393306.1 ENST00000565123.1 |
OSGIN1 |
oxidative stress induced growth inhibitor 1 |
| chr7_-_7680601 | 0.01 |
ENST00000396682.2 |
RPA3 |
replication protein A3, 14kDa |
| chr15_-_50558223 | 0.01 |
ENST00000267845.3 |
HDC |
histidine decarboxylase |
| chr12_-_371994 | 0.01 |
ENST00000343164.4 ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13 |
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr17_+_33914276 | 0.01 |
ENST00000592545.1 ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr11_+_67798363 | 0.01 |
ENST00000525628.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr4_-_90756769 | 0.01 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr6_-_144385698 | 0.01 |
ENST00000444202.1 ENST00000437412.1 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |