Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
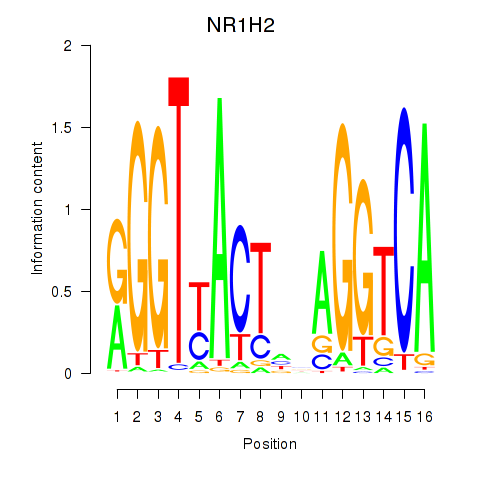
Results for NR1H2
Z-value: 0.85
Transcription factors associated with NR1H2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1H2
|
ENSG00000131408.9 | NR1H2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1H2 | hg19_v2_chr19_+_50879705_50879775 | 0.32 | 4.3e-01 | Click! |
Activity profile of NR1H2 motif
Sorted Z-values of NR1H2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1H2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91573132 | 0.83 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr16_+_53133070 | 0.50 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr17_-_19648916 | 0.44 |
ENST00000444455.1 ENST00000439102.2 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr3_-_195310802 | 0.36 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
| chr1_+_201755452 | 0.33 |
ENST00000438083.1 |
NAV1 |
neuron navigator 1 |
| chr20_+_33292068 | 0.30 |
ENST00000374810.3 ENST00000374809.2 ENST00000451665.1 |
TP53INP2 |
tumor protein p53 inducible nuclear protein 2 |
| chr12_-_91573249 | 0.29 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr20_+_34680620 | 0.29 |
ENST00000430276.1 ENST00000373950.2 ENST00000452261.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr1_+_6615241 | 0.28 |
ENST00000333172.6 ENST00000328191.4 ENST00000351136.3 |
TAS1R1 |
taste receptor, type 1, member 1 |
| chr4_-_39640700 | 0.27 |
ENST00000295958.5 |
SMIM14 |
small integral membrane protein 14 |
| chr4_-_142134031 | 0.25 |
ENST00000420921.2 |
RNF150 |
ring finger protein 150 |
| chr4_-_681114 | 0.23 |
ENST00000503156.1 |
MFSD7 |
major facilitator superfamily domain containing 7 |
| chr20_-_39928705 | 0.22 |
ENST00000436099.2 ENST00000309060.3 ENST00000373261.1 ENST00000436440.2 ENST00000540170.1 ENST00000557816.1 ENST00000560361.1 |
ZHX3 |
zinc fingers and homeoboxes 3 |
| chr1_+_23037323 | 0.22 |
ENST00000544305.1 ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2 |
EPH receptor B2 |
| chr7_-_56160666 | 0.20 |
ENST00000297373.2 |
PHKG1 |
phosphorylase kinase, gamma 1 (muscle) |
| chr7_-_56160625 | 0.20 |
ENST00000446428.1 ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1 |
phosphorylase kinase, gamma 1 (muscle) |
| chr8_+_27183033 | 0.20 |
ENST00000420218.2 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr7_-_138363824 | 0.19 |
ENST00000419765.3 |
SVOPL |
SVOP-like |
| chr8_+_27182862 | 0.19 |
ENST00000521164.1 ENST00000346049.5 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr11_+_10326612 | 0.18 |
ENST00000534464.1 ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM |
adrenomedullin |
| chr15_+_96875657 | 0.17 |
ENST00000559679.1 ENST00000394171.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr22_-_24096562 | 0.17 |
ENST00000398465.3 |
VPREB3 |
pre-B lymphocyte 3 |
| chr22_+_30792846 | 0.17 |
ENST00000312932.9 ENST00000428195.1 |
SEC14L2 |
SEC14-like 2 (S. cerevisiae) |
| chr15_+_75498739 | 0.16 |
ENST00000565074.1 |
C15orf39 |
chromosome 15 open reading frame 39 |
| chr20_-_39928756 | 0.16 |
ENST00000432768.2 |
ZHX3 |
zinc fingers and homeoboxes 3 |
| chr22_-_24096630 | 0.15 |
ENST00000248948.3 |
VPREB3 |
pre-B lymphocyte 3 |
| chr12_-_56122761 | 0.15 |
ENST00000552164.1 ENST00000420846.3 ENST00000257857.4 |
CD63 |
CD63 molecule |
| chrX_+_21874105 | 0.15 |
ENST00000429584.2 |
YY2 |
YY2 transcription factor |
| chr11_+_65190245 | 0.15 |
ENST00000499732.1 ENST00000501122.2 ENST00000601801.1 |
NEAT1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr13_-_33112823 | 0.15 |
ENST00000504114.1 |
N4BP2L2 |
NEDD4 binding protein 2-like 2 |
| chr19_-_46285646 | 0.15 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr12_+_56511943 | 0.15 |
ENST00000257940.2 ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10 ESYT1 |
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr5_-_133968529 | 0.15 |
ENST00000402673.2 |
SAR1B |
SAR1 homolog B (S. cerevisiae) |
| chr3_-_51994694 | 0.14 |
ENST00000395014.2 |
PCBP4 |
poly(rC) binding protein 4 |
| chr10_+_1095416 | 0.13 |
ENST00000358220.1 |
WDR37 |
WD repeat domain 37 |
| chr11_+_77774897 | 0.13 |
ENST00000281030.2 |
THRSP |
thyroid hormone responsive |
| chr4_-_7941596 | 0.13 |
ENST00000420658.1 ENST00000358461.2 |
AFAP1 |
actin filament associated protein 1 |
| chr10_+_81838411 | 0.13 |
ENST00000372281.3 ENST00000372277.3 ENST00000372275.1 ENST00000372274.1 |
TMEM254 |
transmembrane protein 254 |
| chr15_+_89010923 | 0.13 |
ENST00000353598.6 |
MRPS11 |
mitochondrial ribosomal protein S11 |
| chr12_-_56123444 | 0.13 |
ENST00000546457.1 ENST00000549117.1 |
CD63 |
CD63 molecule |
| chr10_-_70287231 | 0.12 |
ENST00000609923.1 |
SLC25A16 |
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chrX_+_52240504 | 0.12 |
ENST00000399805.2 |
XAGE1B |
X antigen family, member 1B |
| chr15_+_96876340 | 0.12 |
ENST00000453270.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr16_+_4674814 | 0.12 |
ENST00000415496.1 ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr6_+_33378738 | 0.12 |
ENST00000374512.3 ENST00000374516.3 |
PHF1 |
PHD finger protein 1 |
| chr13_+_49684445 | 0.12 |
ENST00000398316.3 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr19_-_45873642 | 0.12 |
ENST00000485403.2 ENST00000586856.1 ENST00000586131.1 ENST00000391940.4 ENST00000221481.6 ENST00000391944.3 ENST00000391945.4 |
ERCC2 |
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr15_-_102285007 | 0.12 |
ENST00000560292.2 |
RP11-89K11.1 |
Uncharacterized protein |
| chr4_-_39033963 | 0.12 |
ENST00000381938.3 |
TMEM156 |
transmembrane protein 156 |
| chr17_-_2169425 | 0.11 |
ENST00000570606.1 ENST00000354901.4 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
| chr20_-_5485166 | 0.11 |
ENST00000589201.1 ENST00000379053.4 |
LINC00654 |
long intergenic non-protein coding RNA 654 |
| chr11_-_7985193 | 0.11 |
ENST00000328600.2 |
NLRP10 |
NLR family, pyrin domain containing 10 |
| chr19_-_38806540 | 0.11 |
ENST00000592694.1 |
YIF1B |
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr11_-_62521614 | 0.11 |
ENST00000527994.1 ENST00000394807.3 |
ZBTB3 |
zinc finger and BTB domain containing 3 |
| chr11_-_68518910 | 0.11 |
ENST00000544963.1 ENST00000443940.2 |
MTL5 |
metallothionein-like 5, testis-specific (tesmin) |
| chr4_+_108910870 | 0.11 |
ENST00000403312.1 ENST00000603302.1 ENST00000309522.3 |
HADH |
hydroxyacyl-CoA dehydrogenase |
| chr19_-_22193731 | 0.11 |
ENST00000601773.1 ENST00000397126.4 ENST00000601993.1 ENST00000599916.1 |
ZNF208 |
zinc finger protein 208 |
| chr16_+_4674787 | 0.11 |
ENST00000262370.7 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr16_-_68034470 | 0.10 |
ENST00000412757.2 |
DPEP2 |
dipeptidase 2 |
| chr16_+_3550924 | 0.10 |
ENST00000576634.1 ENST00000574369.1 ENST00000341633.5 ENST00000417763.2 ENST00000571025.1 |
CLUAP1 |
clusterin associated protein 1 |
| chr9_-_130829588 | 0.10 |
ENST00000373078.4 |
NAIF1 |
nuclear apoptosis inducing factor 1 |
| chr11_+_67171391 | 0.10 |
ENST00000312390.5 |
TBC1D10C |
TBC1 domain family, member 10C |
| chr6_-_167571817 | 0.10 |
ENST00000366834.1 |
GPR31 |
G protein-coupled receptor 31 |
| chr14_-_94970559 | 0.10 |
ENST00000556881.1 |
SERPINA12 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr11_-_68519026 | 0.09 |
ENST00000255087.5 |
MTL5 |
metallothionein-like 5, testis-specific (tesmin) |
| chr7_+_2687173 | 0.09 |
ENST00000403167.1 |
TTYH3 |
tweety family member 3 |
| chr17_-_8198636 | 0.09 |
ENST00000577745.1 ENST00000579192.1 ENST00000396278.1 |
SLC25A35 |
solute carrier family 25, member 35 |
| chr11_+_66276550 | 0.09 |
ENST00000419755.3 |
CTD-3074O7.11 |
Bardet-Biedl syndrome 1 protein |
| chr11_+_67171358 | 0.09 |
ENST00000526387.1 |
TBC1D10C |
TBC1 domain family, member 10C |
| chr14_-_93673353 | 0.09 |
ENST00000556566.1 ENST00000306954.4 |
C14orf142 |
chromosome 14 open reading frame 142 |
| chr17_-_42580738 | 0.09 |
ENST00000585614.1 ENST00000591680.1 ENST00000434000.1 ENST00000588554.1 ENST00000592154.1 |
GPATCH8 |
G patch domain containing 8 |
| chr4_-_39034542 | 0.09 |
ENST00000344606.6 |
TMEM156 |
transmembrane protein 156 |
| chr14_-_102976135 | 0.09 |
ENST00000560748.1 |
ANKRD9 |
ankyrin repeat domain 9 |
| chr2_-_3521518 | 0.09 |
ENST00000382093.5 |
ADI1 |
acireductone dioxygenase 1 |
| chr16_-_18468926 | 0.09 |
ENST00000545114.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr19_-_38806390 | 0.08 |
ENST00000589247.1 ENST00000329420.8 ENST00000591784.1 |
YIF1B |
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr1_-_9714644 | 0.08 |
ENST00000377320.3 |
C1orf200 |
chromosome 1 open reading frame 200 |
| chr17_-_49198216 | 0.08 |
ENST00000262013.7 ENST00000357122.4 |
SPAG9 |
sperm associated antigen 9 |
| chr14_-_102976091 | 0.08 |
ENST00000286918.4 |
ANKRD9 |
ankyrin repeat domain 9 |
| chr2_+_44502597 | 0.08 |
ENST00000260649.6 ENST00000409387.1 |
SLC3A1 |
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr11_+_64692143 | 0.08 |
ENST00000164133.2 ENST00000532850.1 |
PPP2R5B |
protein phosphatase 2, regulatory subunit B', beta |
| chr5_+_43120985 | 0.08 |
ENST00000515326.1 |
ZNF131 |
zinc finger protein 131 |
| chr15_+_83776137 | 0.08 |
ENST00000322019.9 |
TM6SF1 |
transmembrane 6 superfamily member 1 |
| chr19_-_38806560 | 0.08 |
ENST00000591755.1 ENST00000337679.8 ENST00000339413.6 |
YIF1B |
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr15_+_74610894 | 0.08 |
ENST00000558821.1 ENST00000268082.4 |
CCDC33 |
coiled-coil domain containing 33 |
| chr12_+_52281744 | 0.08 |
ENST00000301190.6 ENST00000538991.1 |
ANKRD33 |
ankyrin repeat domain 33 |
| chr6_-_2972075 | 0.08 |
ENST00000335686.5 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr19_+_41092680 | 0.08 |
ENST00000594298.1 ENST00000597396.1 |
SHKBP1 |
SH3KBP1 binding protein 1 |
| chr20_-_34117447 | 0.08 |
ENST00000246199.2 ENST00000424444.1 ENST00000374345.4 ENST00000444723.1 |
C20orf173 |
chromosome 20 open reading frame 173 |
| chr19_+_57631411 | 0.08 |
ENST00000254181.4 ENST00000600940.1 |
USP29 |
ubiquitin specific peptidase 29 |
| chr19_+_58361185 | 0.08 |
ENST00000339656.5 ENST00000423137.1 |
ZNF587 |
zinc finger protein 587 |
| chr20_-_48747662 | 0.08 |
ENST00000371656.2 |
TMEM189 |
transmembrane protein 189 |
| chr12_+_123944070 | 0.08 |
ENST00000412157.2 |
SNRNP35 |
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr3_-_195938256 | 0.08 |
ENST00000296326.3 |
ZDHHC19 |
zinc finger, DHHC-type containing 19 |
| chr19_-_54850417 | 0.07 |
ENST00000291759.4 |
LILRA4 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4 |
| chr20_+_5987890 | 0.07 |
ENST00000378868.4 |
CRLS1 |
cardiolipin synthase 1 |
| chr1_-_237167718 | 0.07 |
ENST00000464121.2 |
MT1HL1 |
metallothionein 1H-like 1 |
| chr1_-_9129895 | 0.07 |
ENST00000473209.1 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr12_+_123942188 | 0.07 |
ENST00000526639.2 |
SNRNP35 |
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr12_+_22778009 | 0.07 |
ENST00000266517.4 ENST00000335148.3 |
ETNK1 |
ethanolamine kinase 1 |
| chr7_-_92219698 | 0.07 |
ENST00000438306.1 ENST00000445716.1 |
FAM133B |
family with sequence similarity 133, member B |
| chr11_+_86748998 | 0.07 |
ENST00000525018.1 ENST00000355734.4 |
TMEM135 |
transmembrane protein 135 |
| chr2_+_108443388 | 0.07 |
ENST00000354986.4 ENST00000408999.3 |
RGPD4 |
RANBP2-like and GRIP domain containing 4 |
| chr1_-_13115578 | 0.07 |
ENST00000414205.2 |
PRAMEF6 |
PRAME family member 6 |
| chr10_-_70287172 | 0.07 |
ENST00000539557.1 |
SLC25A16 |
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr12_-_120765565 | 0.06 |
ENST00000423423.3 ENST00000308366.4 |
PLA2G1B |
phospholipase A2, group IB (pancreas) |
| chr17_-_76183111 | 0.06 |
ENST00000405273.1 ENST00000590862.1 ENST00000590430.1 ENST00000586613.1 |
TK1 |
thymidine kinase 1, soluble |
| chr2_+_223725652 | 0.06 |
ENST00000357430.3 ENST00000392066.3 |
ACSL3 |
acyl-CoA synthetase long-chain family member 3 |
| chr2_-_107084826 | 0.06 |
ENST00000304514.7 ENST00000409886.3 |
RGPD3 |
RANBP2-like and GRIP domain containing 3 |
| chr16_+_29973351 | 0.06 |
ENST00000602948.1 ENST00000279396.6 ENST00000575829.2 ENST00000561899.2 |
TMEM219 |
transmembrane protein 219 |
| chr11_+_844406 | 0.06 |
ENST00000397404.1 |
TSPAN4 |
tetraspanin 4 |
| chr17_+_42977122 | 0.06 |
ENST00000412523.2 ENST00000331733.4 ENST00000417826.2 |
FAM187A CCDC103 |
family with sequence similarity 187, member A coiled-coil domain containing 103 |
| chrX_+_102631248 | 0.06 |
ENST00000361298.4 ENST00000372645.3 ENST00000372635.1 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr3_+_9691117 | 0.06 |
ENST00000353332.5 ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14 |
myotubularin related protein 14 |
| chr1_-_20126365 | 0.06 |
ENST00000294543.6 ENST00000375122.2 |
TMCO4 |
transmembrane and coiled-coil domains 4 |
| chr15_-_83837983 | 0.06 |
ENST00000562702.1 |
HDGFRP3 |
Hepatoma-derived growth factor-related protein 3 |
| chr19_+_2977444 | 0.06 |
ENST00000246112.4 ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6 |
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr12_+_120972606 | 0.05 |
ENST00000413266.2 |
RNF10 |
ring finger protein 10 |
| chr1_+_28879588 | 0.05 |
ENST00000373830.3 |
TRNAU1AP |
tRNA selenocysteine 1 associated protein 1 |
| chr3_+_39371255 | 0.05 |
ENST00000414803.1 ENST00000545843.1 |
CCR8 |
chemokine (C-C motif) receptor 8 |
| chr20_-_32262165 | 0.05 |
ENST00000606690.1 ENST00000246190.6 ENST00000439478.1 ENST00000375238.4 |
NECAB3 |
N-terminal EF-hand calcium binding protein 3 |
| chr15_-_89010607 | 0.05 |
ENST00000312475.4 |
MRPL46 |
mitochondrial ribosomal protein L46 |
| chr17_+_34087888 | 0.05 |
ENST00000586491.1 ENST00000588628.1 ENST00000285023.4 |
C17orf50 |
chromosome 17 open reading frame 50 |
| chr16_+_21312170 | 0.05 |
ENST00000338573.5 ENST00000561968.1 |
CRYM-AS1 |
CRYM antisense RNA 1 |
| chr8_-_27941380 | 0.05 |
ENST00000413272.2 ENST00000341513.6 |
NUGGC |
nuclear GTPase, germinal center associated |
| chr3_-_49466686 | 0.05 |
ENST00000273598.3 ENST00000436744.2 |
NICN1 |
nicolin 1 |
| chr10_-_28623368 | 0.05 |
ENST00000441595.2 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr2_+_228029281 | 0.05 |
ENST00000396578.3 |
COL4A3 |
collagen, type IV, alpha 3 (Goodpasture antigen) |
| chr11_-_82611448 | 0.05 |
ENST00000393399.2 ENST00000313010.3 |
PRCP |
prolylcarboxypeptidase (angiotensinase C) |
| chr2_+_71357744 | 0.05 |
ENST00000498451.2 |
MPHOSPH10 |
M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) |
| chr17_+_66243715 | 0.05 |
ENST00000359904.3 |
AMZ2 |
archaelysin family metallopeptidase 2 |
| chr19_+_33622996 | 0.04 |
ENST00000592765.1 ENST00000361680.2 ENST00000355868.3 |
WDR88 |
WD repeat domain 88 |
| chr5_+_172410757 | 0.04 |
ENST00000519374.1 ENST00000519911.1 ENST00000265093.4 ENST00000517669.1 |
ATP6V0E1 |
ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 |
| chr1_+_10459111 | 0.04 |
ENST00000541529.1 ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD |
phosphogluconate dehydrogenase |
| chr1_+_36335351 | 0.04 |
ENST00000373206.1 |
AGO1 |
argonaute RISC catalytic component 1 |
| chrX_-_107334750 | 0.04 |
ENST00000340200.5 ENST00000372296.1 ENST00000372295.1 ENST00000361815.5 |
PSMD10 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr6_+_30687978 | 0.04 |
ENST00000327892.8 ENST00000435534.1 |
TUBB |
tubulin, beta class I |
| chr2_-_216946500 | 0.04 |
ENST00000265322.7 |
PECR |
peroxisomal trans-2-enoyl-CoA reductase |
| chr17_+_76142434 | 0.04 |
ENST00000340363.5 ENST00000586999.1 |
C17orf99 |
chromosome 17 open reading frame 99 |
| chr2_+_101179152 | 0.04 |
ENST00000264254.6 |
PDCL3 |
phosducin-like 3 |
| chrX_-_9001116 | 0.04 |
ENST00000428477.1 |
FAM9B |
family with sequence similarity 9, member B |
| chr17_-_15370915 | 0.04 |
ENST00000312177.6 |
CDRT4 |
CMT1A duplicated region transcript 4 |
| chr19_+_39881951 | 0.04 |
ENST00000315588.5 ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29 |
mediator complex subunit 29 |
| chr16_+_88704978 | 0.04 |
ENST00000244241.4 |
IL17C |
interleukin 17C |
| chr17_+_71229346 | 0.03 |
ENST00000535032.2 ENST00000582793.1 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chr12_+_52282001 | 0.03 |
ENST00000340970.4 |
ANKRD33 |
ankyrin repeat domain 33 |
| chr17_+_27047570 | 0.03 |
ENST00000472628.1 ENST00000578181.1 |
RPL23A |
ribosomal protein L23a |
| chr16_-_22012419 | 0.03 |
ENST00000537222.2 ENST00000424898.2 ENST00000286143.6 |
PDZD9 |
PDZ domain containing 9 |
| chrX_-_107334790 | 0.03 |
ENST00000217958.3 |
PSMD10 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr19_+_41768401 | 0.03 |
ENST00000352456.3 ENST00000595018.1 ENST00000597725.1 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr6_-_144385698 | 0.03 |
ENST00000444202.1 ENST00000437412.1 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
| chr7_+_153584166 | 0.03 |
ENST00000404039.1 |
DPP6 |
dipeptidyl-peptidase 6 |
| chr20_+_57556263 | 0.03 |
ENST00000602795.1 ENST00000344018.3 |
NELFCD |
negative elongation factor complex member C/D |
| chr5_+_446253 | 0.03 |
ENST00000315013.5 |
EXOC3 |
exocyst complex component 3 |
| chr20_+_35918035 | 0.03 |
ENST00000373606.3 ENST00000397156.3 ENST00000397150.1 ENST00000397152.3 |
MANBAL |
mannosidase, beta A, lysosomal-like |
| chr19_+_16254488 | 0.03 |
ENST00000588246.1 ENST00000593031.1 |
HSH2D |
hematopoietic SH2 domain containing |
| chr3_+_39371191 | 0.03 |
ENST00000326306.4 |
CCR8 |
chemokine (C-C motif) receptor 8 |
| chr16_+_56995762 | 0.03 |
ENST00000200676.3 ENST00000379780.2 |
CETP |
cholesteryl ester transfer protein, plasma |
| chr1_+_100818156 | 0.03 |
ENST00000336454.3 |
CDC14A |
cell division cycle 14A |
| chr7_+_45067265 | 0.03 |
ENST00000474617.1 |
CCM2 |
cerebral cavernous malformation 2 |
| chr7_+_56131917 | 0.03 |
ENST00000434526.2 ENST00000275607.9 ENST00000395435.2 ENST00000413952.2 ENST00000342190.6 ENST00000437307.2 ENST00000413756.1 ENST00000451338.1 |
SUMF2 |
sulfatase modifying factor 2 |
| chr2_-_32390801 | 0.03 |
ENST00000608489.1 |
RP11-563N4.1 |
RP11-563N4.1 |
| chr1_-_153949751 | 0.03 |
ENST00000428469.1 |
JTB |
jumping translocation breakpoint |
| chr1_+_87595497 | 0.03 |
ENST00000471417.1 |
RP5-1052I5.1 |
long intergenic non-protein coding RNA 1140 |
| chr16_+_56995854 | 0.03 |
ENST00000566128.1 |
CETP |
cholesteryl ester transfer protein, plasma |
| chr17_-_62097904 | 0.03 |
ENST00000583366.1 |
ICAM2 |
intercellular adhesion molecule 2 |
| chr8_+_74903580 | 0.03 |
ENST00000284818.2 ENST00000518893.1 |
LY96 |
lymphocyte antigen 96 |
| chr7_-_73153122 | 0.03 |
ENST00000458339.1 |
ABHD11 |
abhydrolase domain containing 11 |
| chr1_+_100818009 | 0.03 |
ENST00000370125.2 ENST00000361544.6 ENST00000370124.3 |
CDC14A |
cell division cycle 14A |
| chr17_-_27054952 | 0.02 |
ENST00000580518.1 |
TLCD1 |
TLC domain containing 1 |
| chr17_-_5026397 | 0.02 |
ENST00000250076.3 |
ZNF232 |
zinc finger protein 232 |
| chr12_-_125398850 | 0.02 |
ENST00000535859.1 ENST00000546271.1 ENST00000540700.1 ENST00000546120.1 |
UBC |
ubiquitin C |
| chr3_-_169587621 | 0.02 |
ENST00000523069.1 ENST00000316428.5 ENST00000264676.5 |
LRRC31 |
leucine rich repeat containing 31 |
| chr5_-_138862326 | 0.02 |
ENST00000330794.4 |
TMEM173 |
transmembrane protein 173 |
| chr7_-_73153161 | 0.02 |
ENST00000395147.4 |
ABHD11 |
abhydrolase domain containing 11 |
| chr12_-_125399573 | 0.02 |
ENST00000339647.5 |
UBC |
ubiquitin C |
| chr1_-_169337176 | 0.02 |
ENST00000472647.1 ENST00000367811.3 |
NME7 |
NME/NM23 family member 7 |
| chr6_+_31674639 | 0.02 |
ENST00000556581.1 ENST00000375832.4 ENST00000503322.1 |
LY6G6F MEGT1 |
lymphocyte antigen 6 complex, locus G6F HCG43720, isoform CRA_a; Lymphocyte antigen 6 complex locus protein G6f; Megakaryocyte-enhanced gene transcript 1 protein; Uncharacterized protein |
| chr4_-_84205905 | 0.02 |
ENST00000311461.7 ENST00000311469.4 ENST00000439031.2 |
COQ2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
| chr9_-_36400213 | 0.02 |
ENST00000259605.6 ENST00000353739.4 |
RNF38 |
ring finger protein 38 |
| chr1_-_161207953 | 0.02 |
ENST00000367982.4 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
| chr11_-_62420757 | 0.02 |
ENST00000330574.2 |
INTS5 |
integrator complex subunit 5 |
| chr7_+_105172532 | 0.02 |
ENST00000257700.2 |
RINT1 |
RAD50 interactor 1 |
| chr1_-_161208013 | 0.02 |
ENST00000515452.1 ENST00000367983.4 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
| chr8_-_93115445 | 0.02 |
ENST00000523629.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr19_-_6670128 | 0.02 |
ENST00000245912.3 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
| chr17_+_32683456 | 0.02 |
ENST00000225844.2 |
CCL13 |
chemokine (C-C motif) ligand 13 |
| chr11_-_506316 | 0.02 |
ENST00000532055.1 ENST00000531540.1 |
RNH1 |
ribonuclease/angiogenin inhibitor 1 |
| chr12_-_7125770 | 0.02 |
ENST00000261407.4 |
LPCAT3 |
lysophosphatidylcholine acyltransferase 3 |
| chr1_-_155829191 | 0.02 |
ENST00000437809.1 |
GON4L |
gon-4-like (C. elegans) |
| chrX_+_107334895 | 0.02 |
ENST00000372232.3 ENST00000345734.3 ENST00000372254.3 |
ATG4A |
autophagy related 4A, cysteine peptidase |
| chr1_-_67142710 | 0.01 |
ENST00000502413.2 |
AL139147.1 |
Uncharacterized protein |
| chr8_+_38065104 | 0.01 |
ENST00000521311.1 |
BAG4 |
BCL2-associated athanogene 4 |
| chr7_+_101460882 | 0.01 |
ENST00000292535.7 ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1 |
cut-like homeobox 1 |
| chr4_-_71705082 | 0.01 |
ENST00000439371.1 ENST00000499044.2 |
GRSF1 |
G-rich RNA sequence binding factor 1 |
| chrX_-_2847366 | 0.01 |
ENST00000381154.1 |
ARSD |
arylsulfatase D |
| chr15_+_64386261 | 0.01 |
ENST00000560829.1 |
SNX1 |
sorting nexin 1 |
| chr5_-_180671172 | 0.01 |
ENST00000512805.1 |
GNB2L1 |
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr16_-_2205352 | 0.01 |
ENST00000563192.1 |
RP11-304L19.5 |
RP11-304L19.5 |
| chr15_+_67835517 | 0.01 |
ENST00000395476.2 |
MAP2K5 |
mitogen-activated protein kinase kinase 5 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.4 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 1.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.3 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.2 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.3 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:1990822 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.0 | GO:0002353 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.4 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:0097319 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.4 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 1.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |