Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for NR1I2
Z-value: 0.69
Transcription factors associated with NR1I2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I2
|
ENSG00000144852.12 | NR1I2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I2 | hg19_v2_chr3_+_119499331_119499331, hg19_v2_chr3_+_119501557_119501557 | 0.18 | 6.6e-01 | Click! |
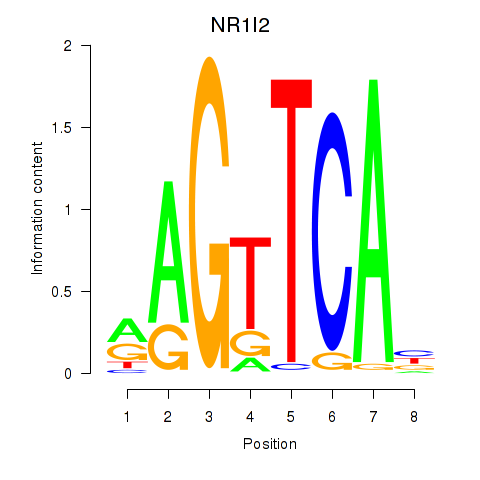
Activity profile of NR1I2 motif
Sorted Z-values of NR1I2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91576429 | 2.08 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr12_-_91576561 | 0.87 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr12_-_56106060 | 0.81 |
ENST00000452168.2 |
ITGA7 |
integrin, alpha 7 |
| chr12_-_91576750 | 0.78 |
ENST00000228329.5 ENST00000303320.3 ENST00000052754.5 |
DCN |
decorin |
| chr10_+_69865866 | 0.77 |
ENST00000354393.2 |
MYPN |
myopalladin |
| chr3_+_157154578 | 0.74 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr6_-_46293378 | 0.74 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr1_+_223101757 | 0.70 |
ENST00000284476.6 |
DISP1 |
dispatched homolog 1 (Drosophila) |
| chr11_-_111783595 | 0.68 |
ENST00000528628.1 |
CRYAB |
crystallin, alpha B |
| chr8_-_120685608 | 0.62 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr12_-_96184533 | 0.62 |
ENST00000343702.4 ENST00000344911.4 |
NTN4 |
netrin 4 |
| chr14_-_90085458 | 0.60 |
ENST00000345097.4 ENST00000555855.1 ENST00000555353.1 |
FOXN3 |
forkhead box N3 |
| chr5_+_149569520 | 0.59 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr6_-_154751629 | 0.56 |
ENST00000424998.1 |
CNKSR3 |
CNKSR family member 3 |
| chr5_+_92919043 | 0.54 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr6_+_132891461 | 0.54 |
ENST00000275198.1 |
TAAR6 |
trace amine associated receptor 6 |
| chr4_-_159080806 | 0.53 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr6_+_84743436 | 0.53 |
ENST00000257776.4 |
MRAP2 |
melanocortin 2 receptor accessory protein 2 |
| chr11_+_117073850 | 0.53 |
ENST00000529622.1 |
TAGLN |
transgelin |
| chr13_+_32605437 | 0.51 |
ENST00000380250.3 |
FRY |
furry homolog (Drosophila) |
| chr3_+_12330560 | 0.51 |
ENST00000397026.2 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr4_+_124317940 | 0.50 |
ENST00000505319.1 ENST00000339241.1 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr3_+_8543393 | 0.49 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr2_+_30454390 | 0.49 |
ENST00000395323.3 ENST00000406087.1 ENST00000404397.1 |
LBH |
limb bud and heart development |
| chr4_-_159094194 | 0.48 |
ENST00000592057.1 ENST00000585682.1 ENST00000393807.5 |
FAM198B |
family with sequence similarity 198, member B |
| chr5_-_158526693 | 0.46 |
ENST00000380654.4 |
EBF1 |
early B-cell factor 1 |
| chr5_-_158526756 | 0.45 |
ENST00000313708.6 ENST00000517373.1 |
EBF1 |
early B-cell factor 1 |
| chr3_-_114343039 | 0.44 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr3_-_9994021 | 0.44 |
ENST00000411976.2 ENST00000412055.1 |
PRRT3 |
proline-rich transmembrane protein 3 |
| chr3_+_8543533 | 0.43 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr8_+_37654424 | 0.43 |
ENST00000315215.7 |
GPR124 |
G protein-coupled receptor 124 |
| chr1_-_144994909 | 0.42 |
ENST00000369347.4 ENST00000369354.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr3_+_8543561 | 0.40 |
ENST00000397386.3 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr11_-_111794446 | 0.38 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr22_-_24641027 | 0.38 |
ENST00000398292.3 ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5 |
gamma-glutamyltransferase 5 |
| chr13_-_33859819 | 0.37 |
ENST00000336934.5 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr6_-_85474219 | 0.36 |
ENST00000369663.5 |
TBX18 |
T-box 18 |
| chr6_+_155537771 | 0.36 |
ENST00000275246.7 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
| chr1_-_144995074 | 0.35 |
ENST00000534536.1 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr1_+_43291220 | 0.35 |
ENST00000372514.3 |
ERMAP |
erythroblast membrane-associated protein (Scianna blood group) |
| chr3_-_49722523 | 0.34 |
ENST00000448220.1 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr8_-_93115445 | 0.33 |
ENST00000523629.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_-_114790179 | 0.32 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chrX_-_100183894 | 0.32 |
ENST00000328526.5 ENST00000372956.2 |
XKRX |
XK, Kell blood group complex subunit-related, X-linked |
| chr3_+_69985734 | 0.31 |
ENST00000314557.6 ENST00000394351.3 |
MITF |
microphthalmia-associated transcription factor |
| chr6_-_167040731 | 0.31 |
ENST00000265678.4 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr16_+_67207838 | 0.31 |
ENST00000566871.1 ENST00000268605.7 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr16_+_67207872 | 0.31 |
ENST00000563258.1 ENST00000568146.1 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr12_-_6665200 | 0.30 |
ENST00000336604.4 ENST00000396840.2 ENST00000356896.4 |
IFFO1 |
intermediate filament family orphan 1 |
| chr10_-_44070016 | 0.30 |
ENST00000374446.2 ENST00000426961.1 ENST00000535642.1 |
ZNF239 |
zinc finger protein 239 |
| chr13_-_33112823 | 0.29 |
ENST00000504114.1 |
N4BP2L2 |
NEDD4 binding protein 2-like 2 |
| chr5_-_131347501 | 0.29 |
ENST00000543479.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr1_-_144995002 | 0.28 |
ENST00000369356.4 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr10_-_33625154 | 0.28 |
ENST00000265371.4 |
NRP1 |
neuropilin 1 |
| chr11_+_64879317 | 0.28 |
ENST00000526809.1 ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2 |
transmembrane 7 superfamily member 2 |
| chrX_+_149531524 | 0.28 |
ENST00000370401.2 |
MAMLD1 |
mastermind-like domain containing 1 |
| chr5_-_124080203 | 0.28 |
ENST00000504926.1 |
ZNF608 |
zinc finger protein 608 |
| chr5_-_124081008 | 0.27 |
ENST00000306315.5 |
ZNF608 |
zinc finger protein 608 |
| chr15_-_81616446 | 0.25 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr7_-_27169801 | 0.25 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr1_+_151739131 | 0.25 |
ENST00000400999.1 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chrX_+_102840408 | 0.24 |
ENST00000468024.1 ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4 |
transcription elongation factor A (SII)-like 4 |
| chr10_-_25010795 | 0.24 |
ENST00000416305.1 ENST00000376410.2 |
ARHGAP21 |
Rho GTPase activating protein 21 |
| chr9_+_17134980 | 0.23 |
ENST00000380647.3 |
CNTLN |
centlein, centrosomal protein |
| chr10_+_135192695 | 0.23 |
ENST00000368539.4 ENST00000278060.5 ENST00000357296.3 |
PAOX |
polyamine oxidase (exo-N4-amino) |
| chr1_+_236305826 | 0.23 |
ENST00000366592.3 ENST00000366591.4 |
GPR137B |
G protein-coupled receptor 137B |
| chr9_-_96215822 | 0.23 |
ENST00000375412.5 |
FAM120AOS |
family with sequence similarity 120A opposite strand |
| chr9_-_73483958 | 0.22 |
ENST00000377101.1 ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr22_-_32341336 | 0.22 |
ENST00000248984.3 |
C22orf24 |
chromosome 22 open reading frame 24 |
| chr2_+_109223595 | 0.22 |
ENST00000410093.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr14_+_88852059 | 0.21 |
ENST00000045347.7 |
SPATA7 |
spermatogenesis associated 7 |
| chr20_+_54933971 | 0.21 |
ENST00000371384.3 ENST00000437418.1 |
FAM210B |
family with sequence similarity 210, member B |
| chr12_+_117176090 | 0.20 |
ENST00000257575.4 ENST00000407967.3 ENST00000392549.2 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr2_+_109237717 | 0.20 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr8_-_13372395 | 0.20 |
ENST00000276297.4 ENST00000511869.1 |
DLC1 |
deleted in liver cancer 1 |
| chr14_-_73493784 | 0.20 |
ENST00000553891.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr17_-_7145106 | 0.20 |
ENST00000577035.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr17_-_9940058 | 0.20 |
ENST00000585266.1 |
GAS7 |
growth arrest-specific 7 |
| chr5_-_127674883 | 0.19 |
ENST00000507835.1 |
FBN2 |
fibrillin 2 |
| chr1_-_144994840 | 0.19 |
ENST00000369351.3 ENST00000369349.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr11_+_118478313 | 0.19 |
ENST00000356063.5 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
| chr6_+_73331918 | 0.19 |
ENST00000402622.2 ENST00000355635.3 ENST00000403813.2 ENST00000414165.2 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr16_+_28875126 | 0.19 |
ENST00000359285.5 ENST00000538342.1 |
SH2B1 |
SH2B adaptor protein 1 |
| chr5_+_137203465 | 0.19 |
ENST00000239926.4 |
MYOT |
myotilin |
| chr2_-_219433014 | 0.19 |
ENST00000418019.1 ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37 |
ubiquitin specific peptidase 37 |
| chr17_-_7531121 | 0.19 |
ENST00000573566.1 ENST00000269298.5 |
SAT2 |
spermidine/spermine N1-acetyltransferase family member 2 |
| chr3_-_178969403 | 0.19 |
ENST00000314235.5 ENST00000392685.2 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr20_-_44485835 | 0.19 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr11_+_308143 | 0.19 |
ENST00000399817.4 |
IFITM2 |
interferon induced transmembrane protein 2 |
| chr9_-_74061751 | 0.18 |
ENST00000357533.2 ENST00000423814.3 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr5_-_133747589 | 0.18 |
ENST00000458198.2 |
CDKN2AIPNL |
CDKN2A interacting protein N-terminal like |
| chr1_+_229440129 | 0.18 |
ENST00000366688.3 |
SPHAR |
S-phase response (cyclin related) |
| chr1_+_218519577 | 0.18 |
ENST00000366930.4 ENST00000366929.4 |
TGFB2 |
transforming growth factor, beta 2 |
| chr9_+_33795533 | 0.18 |
ENST00000379405.3 |
PRSS3 |
protease, serine, 3 |
| chr14_-_73493825 | 0.18 |
ENST00000318876.5 ENST00000556143.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr12_-_120884175 | 0.18 |
ENST00000546954.1 |
TRIAP1 |
TP53 regulated inhibitor of apoptosis 1 |
| chr17_-_14683517 | 0.17 |
ENST00000379640.1 |
AC005863.1 |
AC005863.1 |
| chr1_+_25944341 | 0.17 |
ENST00000263979.3 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
| chr12_+_58013693 | 0.17 |
ENST00000320442.4 ENST00000379218.2 |
SLC26A10 |
solute carrier family 26, member 10 |
| chr17_-_73761222 | 0.17 |
ENST00000437911.1 ENST00000225614.2 |
GALK1 |
galactokinase 1 |
| chr14_+_51026743 | 0.17 |
ENST00000358385.6 ENST00000357032.3 ENST00000354525.4 |
ATL1 |
atlastin GTPase 1 |
| chr11_+_308217 | 0.17 |
ENST00000602569.1 |
IFITM2 |
interferon induced transmembrane protein 2 |
| chr11_+_108093559 | 0.17 |
ENST00000278616.4 |
ATM |
ataxia telangiectasia mutated |
| chr15_+_43809797 | 0.17 |
ENST00000399453.1 ENST00000300231.5 |
MAP1A |
microtubule-associated protein 1A |
| chr14_-_76447494 | 0.16 |
ENST00000238682.3 |
TGFB3 |
transforming growth factor, beta 3 |
| chr5_+_134074231 | 0.16 |
ENST00000514518.1 |
CAMLG |
calcium modulating ligand |
| chr6_+_126102292 | 0.16 |
ENST00000368357.3 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr7_+_150065879 | 0.16 |
ENST00000397281.2 ENST00000444957.1 ENST00000466559.1 ENST00000489432.2 ENST00000475514.1 ENST00000482680.1 ENST00000488943.1 ENST00000518514.1 ENST00000478789.1 |
REPIN1 ZNF775 |
replication initiator 1 zinc finger protein 775 |
| chr1_+_160097462 | 0.16 |
ENST00000447527.1 |
ATP1A2 |
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr13_+_115000556 | 0.15 |
ENST00000252458.6 |
CDC16 |
cell division cycle 16 |
| chrX_+_135279179 | 0.15 |
ENST00000370676.3 |
FHL1 |
four and a half LIM domains 1 |
| chr5_+_140810132 | 0.15 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr11_-_66445219 | 0.15 |
ENST00000525754.1 ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B |
RNA binding motif protein 4B |
| chr16_-_28937027 | 0.15 |
ENST00000358201.4 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr9_+_35829208 | 0.15 |
ENST00000439587.2 ENST00000377991.4 |
TMEM8B |
transmembrane protein 8B |
| chr1_+_43148625 | 0.15 |
ENST00000436427.1 |
YBX1 |
Y box binding protein 1 |
| chr11_+_120973375 | 0.14 |
ENST00000264037.2 |
TECTA |
tectorin alpha |
| chr13_+_115000339 | 0.14 |
ENST00000360383.3 ENST00000375312.3 |
CDC16 |
cell division cycle 16 |
| chr3_+_183894566 | 0.14 |
ENST00000439647.1 |
AP2M1 |
adaptor-related protein complex 2, mu 1 subunit |
| chrX_+_135278908 | 0.14 |
ENST00000539015.1 ENST00000370683.1 |
FHL1 |
four and a half LIM domains 1 |
| chr3_+_69985792 | 0.14 |
ENST00000531774.1 |
MITF |
microphthalmia-associated transcription factor |
| chr4_+_5712898 | 0.14 |
ENST00000264956.6 ENST00000382674.2 |
EVC |
Ellis van Creveld syndrome |
| chr19_+_2977444 | 0.14 |
ENST00000246112.4 ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6 |
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chrX_+_16668278 | 0.14 |
ENST00000380200.3 |
S100G |
S100 calcium binding protein G |
| chr1_+_196788887 | 0.14 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr10_+_76586348 | 0.14 |
ENST00000372724.1 ENST00000287239.4 ENST00000372714.1 |
KAT6B |
K(lysine) acetyltransferase 6B |
| chr3_+_137906109 | 0.13 |
ENST00000481646.1 ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr19_-_43690674 | 0.13 |
ENST00000342951.6 ENST00000366175.3 |
PSG5 |
pregnancy specific beta-1-glycoprotein 5 |
| chr10_+_135192782 | 0.13 |
ENST00000480071.2 |
PAOX |
polyamine oxidase (exo-N4-amino) |
| chr7_+_22766766 | 0.13 |
ENST00000426291.1 ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6 |
interleukin 6 (interferon, beta 2) |
| chr1_+_74701062 | 0.13 |
ENST00000326637.3 |
TNNI3K |
TNNI3 interacting kinase |
| chrX_-_71525742 | 0.13 |
ENST00000450875.1 ENST00000417400.1 ENST00000431381.1 ENST00000445983.1 |
CITED1 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chrX_-_72299258 | 0.13 |
ENST00000453389.1 ENST00000373519.1 |
PABPC1L2A |
poly(A) binding protein, cytoplasmic 1-like 2A |
| chr15_+_96875657 | 0.13 |
ENST00000559679.1 ENST00000394171.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr3_+_13978886 | 0.13 |
ENST00000524375.1 ENST00000326972.8 |
TPRXL |
tetra-peptide repeat homeobox-like |
| chr3_+_37284824 | 0.13 |
ENST00000431105.1 |
GOLGA4 |
golgin A4 |
| chr19_+_4153598 | 0.13 |
ENST00000078445.2 ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3 |
cAMP responsive element binding protein 3-like 3 |
| chr2_-_65593784 | 0.13 |
ENST00000443619.2 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
| chr9_-_130829588 | 0.13 |
ENST00000373078.4 |
NAIF1 |
nuclear apoptosis inducing factor 1 |
| chrX_+_72223352 | 0.13 |
ENST00000373521.2 ENST00000538388.1 |
PABPC1L2B |
poly(A) binding protein, cytoplasmic 1-like 2B |
| chr3_+_46618727 | 0.13 |
ENST00000296145.5 |
TDGF1 |
teratocarcinoma-derived growth factor 1 |
| chr7_+_97910962 | 0.13 |
ENST00000539286.1 |
BRI3 |
brain protein I3 |
| chr19_+_41117770 | 0.13 |
ENST00000601032.1 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr1_-_182640988 | 0.13 |
ENST00000367556.1 |
RGS8 |
regulator of G-protein signaling 8 |
| chr13_+_115000521 | 0.12 |
ENST00000252457.5 ENST00000375308.1 |
CDC16 |
cell division cycle 16 |
| chr12_+_117176113 | 0.12 |
ENST00000319176.7 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr22_+_30163340 | 0.12 |
ENST00000330029.6 ENST00000401406.3 |
UQCR10 |
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr9_+_37650945 | 0.12 |
ENST00000377765.3 |
FRMPD1 |
FERM and PDZ domain containing 1 |
| chr16_+_47495201 | 0.12 |
ENST00000566044.1 ENST00000455779.1 |
PHKB |
phosphorylase kinase, beta |
| chr19_-_46285646 | 0.12 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr7_-_14028488 | 0.12 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chrX_+_152990302 | 0.11 |
ENST00000218104.3 |
ABCD1 |
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr2_-_220083076 | 0.11 |
ENST00000295750.4 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr17_+_75137460 | 0.11 |
ENST00000587820.1 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
| chr16_+_47495225 | 0.11 |
ENST00000299167.8 ENST00000323584.5 ENST00000563376.1 |
PHKB |
phosphorylase kinase, beta |
| chr10_+_116581503 | 0.11 |
ENST00000369248.4 ENST00000369250.3 ENST00000369246.1 |
FAM160B1 |
family with sequence similarity 160, member B1 |
| chr5_-_73937244 | 0.11 |
ENST00000302351.4 ENST00000510316.1 ENST00000508331.1 |
ENC1 |
ectodermal-neural cortex 1 (with BTB domain) |
| chr2_+_10560147 | 0.11 |
ENST00000422133.1 |
HPCAL1 |
hippocalcin-like 1 |
| chr7_+_97910981 | 0.11 |
ENST00000297290.3 |
BRI3 |
brain protein I3 |
| chr11_-_7985193 | 0.11 |
ENST00000328600.2 |
NLRP10 |
NLR family, pyrin domain containing 10 |
| chr12_-_49582978 | 0.11 |
ENST00000301071.7 |
TUBA1A |
tubulin, alpha 1a |
| chr10_-_121296045 | 0.10 |
ENST00000392865.1 |
RGS10 |
regulator of G-protein signaling 10 |
| chr8_+_67579807 | 0.10 |
ENST00000519289.1 ENST00000519561.1 ENST00000521889.1 |
C8orf44-SGK3 C8orf44 |
C8orf44-SGK3 readthrough chromosome 8 open reading frame 44 |
| chr13_+_115000384 | 0.10 |
ENST00000356221.3 ENST00000375310.1 |
CDC16 |
cell division cycle 16 |
| chr5_+_134074191 | 0.10 |
ENST00000297156.2 |
CAMLG |
calcium modulating ligand |
| chr12_+_56511943 | 0.10 |
ENST00000257940.2 ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10 ESYT1 |
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr16_-_67281413 | 0.10 |
ENST00000258201.4 |
FHOD1 |
formin homology 2 domain containing 1 |
| chr20_+_44486246 | 0.10 |
ENST00000255152.2 ENST00000454862.2 |
ZSWIM3 |
zinc finger, SWIM-type containing 3 |
| chr16_-_28936493 | 0.10 |
ENST00000544477.1 ENST00000357573.6 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr7_+_134576151 | 0.10 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr19_-_52511334 | 0.10 |
ENST00000602063.1 ENST00000597747.1 ENST00000594083.1 ENST00000593650.1 ENST00000599631.1 ENST00000598071.1 ENST00000601178.1 ENST00000376716.5 ENST00000391795.3 |
ZNF615 |
zinc finger protein 615 |
| chr2_-_70475701 | 0.10 |
ENST00000282574.4 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr17_-_7082861 | 0.10 |
ENST00000269299.3 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr11_-_627143 | 0.10 |
ENST00000176195.3 |
SCT |
secretin |
| chr17_-_34344991 | 0.10 |
ENST00000591423.1 |
CCL23 |
chemokine (C-C motif) ligand 23 |
| chr19_-_39322497 | 0.10 |
ENST00000221418.4 |
ECH1 |
enoyl CoA hydratase 1, peroxisomal |
| chr11_+_120894781 | 0.10 |
ENST00000529397.1 ENST00000528512.1 ENST00000422003.2 |
TBCEL |
tubulin folding cofactor E-like |
| chr1_-_149858227 | 0.10 |
ENST00000369155.2 |
HIST2H2BE |
histone cluster 2, H2be |
| chr17_-_9479128 | 0.10 |
ENST00000574431.1 |
STX8 |
syntaxin 8 |
| chr1_+_207277590 | 0.09 |
ENST00000367070.3 |
C4BPA |
complement component 4 binding protein, alpha |
| chr20_+_57875658 | 0.09 |
ENST00000371025.3 |
EDN3 |
endothelin 3 |
| chr11_-_63993690 | 0.09 |
ENST00000394546.2 ENST00000541278.1 |
TRPT1 |
tRNA phosphotransferase 1 |
| chr11_-_108369101 | 0.09 |
ENST00000323468.5 |
KDELC2 |
KDEL (Lys-Asp-Glu-Leu) containing 2 |
| chr7_-_131241361 | 0.09 |
ENST00000378555.3 ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL |
podocalyxin-like |
| chr1_-_47407111 | 0.09 |
ENST00000371904.4 |
CYP4A11 |
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr1_-_151965048 | 0.09 |
ENST00000368809.1 |
S100A10 |
S100 calcium binding protein A10 |
| chr14_+_21498666 | 0.09 |
ENST00000481535.1 |
TPPP2 |
tubulin polymerization-promoting protein family member 2 |
| chr9_-_115983641 | 0.09 |
ENST00000238256.3 |
FKBP15 |
FK506 binding protein 15, 133kDa |
| chr10_+_94833642 | 0.09 |
ENST00000224356.4 ENST00000394139.1 |
CYP26A1 |
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr4_+_26322409 | 0.09 |
ENST00000514807.1 ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr19_-_12845550 | 0.09 |
ENST00000242784.4 |
C19orf43 |
chromosome 19 open reading frame 43 |
| chr14_+_55034599 | 0.09 |
ENST00000392067.3 ENST00000357634.3 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr2_-_86790472 | 0.09 |
ENST00000409727.1 |
CHMP3 |
charged multivesicular body protein 3 |
| chr10_-_115904361 | 0.09 |
ENST00000428953.1 ENST00000543782.1 |
C10orf118 |
chromosome 10 open reading frame 118 |
| chr20_+_57875457 | 0.09 |
ENST00000337938.2 ENST00000311585.7 ENST00000371028.2 |
EDN3 |
endothelin 3 |
| chr17_+_33448593 | 0.09 |
ENST00000158009.5 |
FNDC8 |
fibronectin type III domain containing 8 |
| chrX_+_135251835 | 0.09 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr12_-_52585765 | 0.09 |
ENST00000313234.5 ENST00000394815.2 |
KRT80 |
keratin 80 |
| chr16_+_83986827 | 0.09 |
ENST00000393306.1 ENST00000565123.1 |
OSGIN1 |
oxidative stress induced growth inhibitor 1 |
| chr16_-_47495170 | 0.08 |
ENST00000320640.6 ENST00000544001.2 |
ITFG1 |
integrin alpha FG-GAP repeat containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.6 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.2 | 0.5 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.1 | 0.6 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.1 | 0.5 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.4 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.1 | 0.7 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.4 | GO:0042704 | uterine wall breakdown(GO:0042704) |
| 0.1 | 0.5 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.1 | 0.6 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.5 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 1.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.3 | GO:0060301 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.2 | GO:1904882 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.2 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.1 | 0.2 | GO:1905068 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.1 | 1.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.3 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.7 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.2 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.6 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.3 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.3 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:2001138 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 | 0.1 | GO:1903509 | glycolipid metabolic process(GO:0006664) liposaccharide metabolic process(GO:1903509) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.4 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.0 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.7 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.0 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.0 | 0.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0036378 | alkaloid catabolic process(GO:0009822) calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.6 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.7 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.0 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 1.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.0 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.2 | 0.5 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.2 | 0.6 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.3 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.4 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 0.6 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.6 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.2 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.7 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 3.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.0 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |