Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for NR2E1
Z-value: 1.85
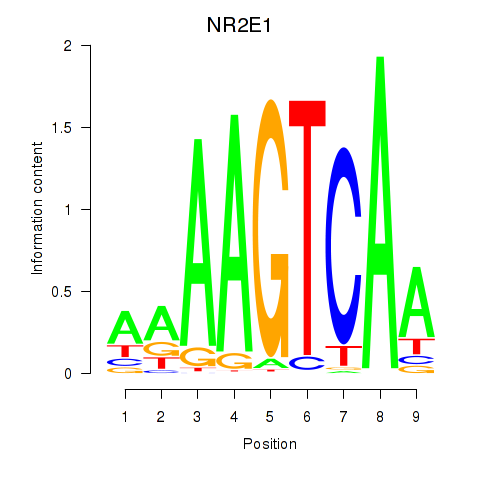
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.7 | NR2E1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E1 | hg19_v2_chr6_+_108487245_108487262 | 0.16 | 7.1e-01 | Click! |
Activity profile of NR2E1 motif
Sorted Z-values of NR2E1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91539918 | 4.25 |
ENST00000548218.1 |
DCN |
decorin |
| chr12_-_91572278 | 2.28 |
ENST00000425043.1 ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN |
decorin |
| chr12_-_91546926 | 1.75 |
ENST00000550758.1 |
DCN |
decorin |
| chr1_+_159141397 | 1.60 |
ENST00000368124.4 ENST00000368125.4 ENST00000416746.1 |
CADM3 |
cell adhesion molecule 3 |
| chr17_-_66951474 | 1.47 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr5_-_9546180 | 1.38 |
ENST00000382496.5 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr1_+_170633047 | 1.34 |
ENST00000239461.6 ENST00000497230.2 |
PRRX1 |
paired related homeobox 1 |
| chr10_-_21463116 | 1.31 |
ENST00000417816.2 |
NEBL |
nebulette |
| chr5_-_150537279 | 1.28 |
ENST00000517486.1 ENST00000377751.5 ENST00000356496.5 ENST00000521512.1 ENST00000517757.1 ENST00000354546.5 |
ANXA6 |
annexin A6 |
| chr10_-_29084886 | 1.21 |
ENST00000608061.1 ENST00000443246.2 ENST00000446012.1 |
LINC00837 |
long intergenic non-protein coding RNA 837 |
| chr8_+_104384616 | 1.18 |
ENST00000520337.1 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr12_-_56236734 | 1.17 |
ENST00000548629.1 |
MMP19 |
matrix metallopeptidase 19 |
| chr10_-_21786179 | 1.16 |
ENST00000377113.5 |
CASC10 |
cancer susceptibility candidate 10 |
| chr5_-_111091948 | 1.15 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr5_+_75699149 | 1.15 |
ENST00000379730.3 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr5_+_75699040 | 1.11 |
ENST00000274364.6 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr3_+_12392971 | 1.09 |
ENST00000287820.6 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr3_+_69788576 | 1.08 |
ENST00000352241.4 ENST00000448226.2 |
MITF |
microphthalmia-associated transcription factor |
| chr3_-_112356944 | 1.06 |
ENST00000461431.1 |
CCDC80 |
coiled-coil domain containing 80 |
| chr12_-_56236690 | 1.03 |
ENST00000322569.4 |
MMP19 |
matrix metallopeptidase 19 |
| chr8_-_122653630 | 0.96 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr9_-_16728161 | 0.93 |
ENST00000603713.1 ENST00000603313.1 |
BNC2 |
basonuclin 2 |
| chr5_+_32711419 | 0.86 |
ENST00000265074.8 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr5_-_90679145 | 0.85 |
ENST00000265138.3 |
ARRDC3 |
arrestin domain containing 3 |
| chr3_+_69985734 | 0.85 |
ENST00000314557.6 ENST00000394351.3 |
MITF |
microphthalmia-associated transcription factor |
| chr2_-_158300556 | 0.84 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr1_+_201708992 | 0.84 |
ENST00000367295.1 |
NAV1 |
neuron navigator 1 |
| chr9_-_95244781 | 0.84 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr5_-_127873659 | 0.82 |
ENST00000262464.4 |
FBN2 |
fibrillin 2 |
| chr3_+_69915385 | 0.82 |
ENST00000314589.5 |
MITF |
microphthalmia-associated transcription factor |
| chr10_+_124320156 | 0.81 |
ENST00000338354.3 ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr8_+_77593448 | 0.79 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr8_+_77593474 | 0.78 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr10_+_124320195 | 0.77 |
ENST00000359586.6 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr2_-_214014959 | 0.73 |
ENST00000442445.1 ENST00000457361.1 ENST00000342002.2 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
| chr3_-_18466026 | 0.69 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr20_-_30310336 | 0.68 |
ENST00000434194.1 ENST00000376062.2 |
BCL2L1 |
BCL2-like 1 |
| chr4_-_157892055 | 0.67 |
ENST00000422544.2 |
PDGFC |
platelet derived growth factor C |
| chr6_-_152958521 | 0.67 |
ENST00000367255.5 ENST00000265368.4 ENST00000448038.1 ENST00000341594.5 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr10_-_106240032 | 0.65 |
ENST00000447860.1 |
RP11-127O4.3 |
RP11-127O4.3 |
| chr4_+_4861385 | 0.63 |
ENST00000382723.4 |
MSX1 |
msh homeobox 1 |
| chr11_-_111794446 | 0.63 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr20_-_30310656 | 0.59 |
ENST00000376055.4 |
BCL2L1 |
BCL2-like 1 |
| chr11_-_63439013 | 0.58 |
ENST00000398868.3 |
ATL3 |
atlastin GTPase 3 |
| chr16_-_73082274 | 0.57 |
ENST00000268489.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr20_-_30310693 | 0.57 |
ENST00000307677.4 ENST00000420653.1 |
BCL2L1 |
BCL2-like 1 |
| chr3_+_69928256 | 0.57 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr12_-_90049828 | 0.55 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr1_+_185703513 | 0.54 |
ENST00000271588.4 ENST00000367492.2 |
HMCN1 |
hemicentin 1 |
| chr3_+_141106458 | 0.53 |
ENST00000509883.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr12_-_90049878 | 0.53 |
ENST00000359142.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_+_79258444 | 0.50 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr11_-_63439174 | 0.49 |
ENST00000332645.4 |
ATL3 |
atlastin GTPase 3 |
| chr1_-_48866517 | 0.48 |
ENST00000371841.1 |
SPATA6 |
spermatogenesis associated 6 |
| chr11_-_111781454 | 0.46 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr9_-_95166841 | 0.46 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr2_+_88047606 | 0.45 |
ENST00000359481.4 |
PLGLB2 |
plasminogen-like B2 |
| chr11_-_111781554 | 0.45 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr5_-_160279207 | 0.45 |
ENST00000327245.5 |
ATP10B |
ATPase, class V, type 10B |
| chr5_+_67588391 | 0.45 |
ENST00000523872.1 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr7_-_38670957 | 0.44 |
ENST00000325590.5 ENST00000428293.2 |
AMPH |
amphiphysin |
| chr7_-_38671098 | 0.44 |
ENST00000356264.2 |
AMPH |
amphiphysin |
| chr9_-_95166884 | 0.43 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr4_+_81187753 | 0.43 |
ENST00000312465.7 ENST00000456523.3 |
FGF5 |
fibroblast growth factor 5 |
| chr12_+_79258547 | 0.43 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr5_+_9546306 | 0.42 |
ENST00000508179.1 |
SNHG18 |
small nucleolar RNA host gene 18 (non-protein coding) |
| chr9_-_16727978 | 0.42 |
ENST00000418777.1 ENST00000468187.2 |
BNC2 |
basonuclin 2 |
| chr17_-_8198636 | 0.42 |
ENST00000577745.1 ENST00000579192.1 ENST00000396278.1 |
SLC25A35 |
solute carrier family 25, member 35 |
| chr7_-_14026123 | 0.41 |
ENST00000420159.2 ENST00000399357.3 ENST00000403527.1 |
ETV1 |
ets variant 1 |
| chr5_+_137203465 | 0.40 |
ENST00000239926.4 |
MYOT |
myotilin |
| chr20_+_43160458 | 0.40 |
ENST00000372889.1 ENST00000372887.1 ENST00000372882.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr13_+_28527647 | 0.39 |
ENST00000567234.1 |
LINC00543 |
long intergenic non-protein coding RNA 543 |
| chr2_+_109237717 | 0.38 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr8_-_116504448 | 0.38 |
ENST00000518018.1 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr6_+_28249332 | 0.37 |
ENST00000259883.3 |
PGBD1 |
piggyBac transposable element derived 1 |
| chr6_+_28249299 | 0.37 |
ENST00000405948.2 |
PGBD1 |
piggyBac transposable element derived 1 |
| chr2_+_110656005 | 0.37 |
ENST00000437679.2 |
LIMS3 |
LIM and senescent cell antigen-like domains 3 |
| chr5_+_118965244 | 0.37 |
ENST00000515256.1 ENST00000509264.1 |
FAM170A |
family with sequence similarity 170, member A |
| chr3_+_69985792 | 0.37 |
ENST00000531774.1 |
MITF |
microphthalmia-associated transcription factor |
| chr1_+_152850787 | 0.37 |
ENST00000368765.3 |
SMCP |
sperm mitochondria-associated cysteine-rich protein |
| chr14_+_51955831 | 0.36 |
ENST00000356218.4 |
FRMD6 |
FERM domain containing 6 |
| chr20_-_30310797 | 0.36 |
ENST00000422920.1 |
BCL2L1 |
BCL2-like 1 |
| chr15_+_83776137 | 0.35 |
ENST00000322019.9 |
TM6SF1 |
transmembrane 6 superfamily member 1 |
| chr5_+_140743859 | 0.35 |
ENST00000518069.1 |
PCDHGA5 |
protocadherin gamma subfamily A, 5 |
| chr8_-_93029520 | 0.34 |
ENST00000521553.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr7_+_116166331 | 0.33 |
ENST00000393468.1 ENST00000393467.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chr1_-_227505289 | 0.32 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr7_-_14026063 | 0.32 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chrX_+_47444613 | 0.32 |
ENST00000445623.1 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
| chr5_+_32711829 | 0.31 |
ENST00000415167.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr5_-_42811986 | 0.31 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr3_+_46616017 | 0.31 |
ENST00000542931.1 |
TDGF1 |
teratocarcinoma-derived growth factor 1 |
| chr7_+_142829162 | 0.31 |
ENST00000291009.3 |
PIP |
prolactin-induced protein |
| chr2_+_131369054 | 0.30 |
ENST00000409602.1 |
POTEJ |
POTE ankyrin domain family, member J |
| chr7_+_70597109 | 0.30 |
ENST00000333538.5 |
WBSCR17 |
Williams-Beuren syndrome chromosome region 17 |
| chr2_-_106013400 | 0.28 |
ENST00000409807.1 |
FHL2 |
four and a half LIM domains 2 |
| chr2_-_85637459 | 0.28 |
ENST00000409921.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr16_+_47495225 | 0.27 |
ENST00000299167.8 ENST00000323584.5 ENST00000563376.1 |
PHKB |
phosphorylase kinase, beta |
| chr7_-_14942944 | 0.27 |
ENST00000403951.2 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr8_-_38126675 | 0.27 |
ENST00000531823.1 ENST00000534339.1 ENST00000524616.1 ENST00000422581.2 ENST00000424479.2 ENST00000419686.2 |
PPAPDC1B |
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr2_+_101437487 | 0.27 |
ENST00000427413.1 ENST00000542504.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr1_-_27240455 | 0.27 |
ENST00000254227.3 |
NR0B2 |
nuclear receptor subfamily 0, group B, member 2 |
| chr7_+_106809406 | 0.27 |
ENST00000468410.1 ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1 |
HMG-box transcription factor 1 |
| chr3_+_141106643 | 0.27 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr7_-_14942283 | 0.26 |
ENST00000402815.1 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr21_-_31744557 | 0.26 |
ENST00000399889.2 |
KRTAP13-2 |
keratin associated protein 13-2 |
| chr16_+_47495201 | 0.26 |
ENST00000566044.1 ENST00000455779.1 |
PHKB |
phosphorylase kinase, beta |
| chr12_+_111471828 | 0.26 |
ENST00000261726.6 |
CUX2 |
cut-like homeobox 2 |
| chr9_-_110251836 | 0.26 |
ENST00000374672.4 |
KLF4 |
Kruppel-like factor 4 (gut) |
| chr1_+_154975258 | 0.26 |
ENST00000417934.2 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
| chr12_-_4754339 | 0.26 |
ENST00000228850.1 |
AKAP3 |
A kinase (PRKA) anchor protein 3 |
| chr8_+_91013676 | 0.26 |
ENST00000519410.1 ENST00000522161.1 ENST00000517761.1 ENST00000520227.1 |
DECR1 |
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr16_+_21689835 | 0.26 |
ENST00000286149.4 ENST00000388958.3 |
OTOA |
otoancorin |
| chr12_+_40618873 | 0.25 |
ENST00000298910.7 |
LRRK2 |
leucine-rich repeat kinase 2 |
| chr1_+_25944341 | 0.25 |
ENST00000263979.3 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
| chr1_-_68698222 | 0.24 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr16_-_66952779 | 0.24 |
ENST00000570262.1 ENST00000394055.3 ENST00000299752.4 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr5_-_88178964 | 0.23 |
ENST00000513252.1 ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr4_+_52917451 | 0.23 |
ENST00000295213.4 ENST00000419395.2 |
SPATA18 |
spermatogenesis associated 18 |
| chr8_+_91013577 | 0.23 |
ENST00000220764.2 |
DECR1 |
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr12_+_64238539 | 0.23 |
ENST00000357825.3 |
SRGAP1 |
SLIT-ROBO Rho GTPase activating protein 1 |
| chr3_-_18466787 | 0.23 |
ENST00000338745.6 ENST00000450898.1 |
SATB1 |
SATB homeobox 1 |
| chr16_-_66952742 | 0.23 |
ENST00000565235.2 ENST00000568632.1 ENST00000565796.1 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr1_-_68698197 | 0.22 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr5_-_88179017 | 0.22 |
ENST00000514028.1 ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr5_-_127873896 | 0.22 |
ENST00000502468.1 |
FBN2 |
fibrillin 2 |
| chr10_+_52152766 | 0.21 |
ENST00000596442.1 |
AC069547.2 |
Uncharacterized protein |
| chr5_+_147774275 | 0.21 |
ENST00000513826.1 |
FBXO38 |
F-box protein 38 |
| chr3_+_138340067 | 0.21 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr14_+_31494841 | 0.21 |
ENST00000556232.1 ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
| chr9_-_99064386 | 0.20 |
ENST00000375262.2 |
HSD17B3 |
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr9_+_2029019 | 0.20 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_-_111291587 | 0.20 |
ENST00000437167.1 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
| chr14_+_31494672 | 0.20 |
ENST00000542754.2 ENST00000313566.6 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
| chrX_-_105282712 | 0.19 |
ENST00000372563.1 |
SERPINA7 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr12_-_82152444 | 0.19 |
ENST00000549325.1 ENST00000550584.2 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr20_+_57875457 | 0.19 |
ENST00000337938.2 ENST00000311585.7 ENST00000371028.2 |
EDN3 |
endothelin 3 |
| chr8_-_16859690 | 0.19 |
ENST00000180166.5 |
FGF20 |
fibroblast growth factor 20 |
| chr2_-_207629997 | 0.19 |
ENST00000454776.2 |
MDH1B |
malate dehydrogenase 1B, NAD (soluble) |
| chr11_-_66104237 | 0.19 |
ENST00000530056.1 |
RIN1 |
Ras and Rab interactor 1 |
| chr6_+_74405501 | 0.18 |
ENST00000437994.2 ENST00000422508.2 |
CD109 |
CD109 molecule |
| chr15_+_23255242 | 0.18 |
ENST00000450802.3 |
GOLGA8I |
golgin A8 family, member I |
| chr12_-_52946923 | 0.18 |
ENST00000267119.5 |
KRT71 |
keratin 71 |
| chr12_+_7169887 | 0.18 |
ENST00000542978.1 |
C1S |
complement component 1, s subcomponent |
| chr1_-_162381907 | 0.18 |
ENST00000367929.2 ENST00000359567.3 |
SH2D1B |
SH2 domain containing 1B |
| chr4_-_52883786 | 0.18 |
ENST00000343457.3 |
LRRC66 |
leucine rich repeat containing 66 |
| chr17_-_40833858 | 0.17 |
ENST00000332438.4 |
CCR10 |
chemokine (C-C motif) receptor 10 |
| chr15_+_31196080 | 0.17 |
ENST00000561607.1 ENST00000565466.1 |
FAN1 |
FANCD2/FANCI-associated nuclease 1 |
| chrX_-_47863348 | 0.17 |
ENST00000376943.3 ENST00000396965.1 ENST00000305127.6 |
ZNF182 |
zinc finger protein 182 |
| chr8_+_24298531 | 0.17 |
ENST00000175238.6 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr8_+_24298597 | 0.17 |
ENST00000380789.1 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr1_-_154155675 | 0.17 |
ENST00000330188.9 ENST00000341485.5 |
TPM3 |
tropomyosin 3 |
| chr6_-_134861089 | 0.16 |
ENST00000606039.1 |
RP11-557H15.4 |
RP11-557H15.4 |
| chr1_+_150229554 | 0.16 |
ENST00000369111.4 |
CA14 |
carbonic anhydrase XIV |
| chr5_-_142780280 | 0.16 |
ENST00000424646.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr6_+_46661575 | 0.16 |
ENST00000450697.1 |
TDRD6 |
tudor domain containing 6 |
| chr12_+_56624436 | 0.16 |
ENST00000266980.4 ENST00000437277.1 |
SLC39A5 |
solute carrier family 39 (zinc transporter), member 5 |
| chr13_-_49975632 | 0.16 |
ENST00000457041.1 ENST00000355854.4 |
CAB39L |
calcium binding protein 39-like |
| chr17_-_73258425 | 0.16 |
ENST00000578348.1 ENST00000582486.1 ENST00000582717.1 |
GGA3 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr4_-_163085141 | 0.16 |
ENST00000427802.2 ENST00000306100.5 |
FSTL5 |
follistatin-like 5 |
| chr11_-_5271122 | 0.16 |
ENST00000330597.3 |
HBG1 |
hemoglobin, gamma A |
| chr7_+_80267973 | 0.16 |
ENST00000394788.3 ENST00000447544.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr2_-_190445499 | 0.15 |
ENST00000261024.2 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr2_-_87248975 | 0.15 |
ENST00000409310.2 ENST00000355705.3 |
PLGLB1 |
plasminogen-like B1 |
| chr19_+_39759154 | 0.15 |
ENST00000331982.5 |
IFNL2 |
interferon, lambda 2 |
| chr16_+_56385290 | 0.15 |
ENST00000564727.1 |
GNAO1 |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chrX_-_65259914 | 0.14 |
ENST00000374737.4 ENST00000455586.2 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr4_-_163085107 | 0.14 |
ENST00000379164.4 |
FSTL5 |
follistatin-like 5 |
| chrX_+_150866779 | 0.14 |
ENST00000370353.3 |
PRRG3 |
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr6_+_161123270 | 0.14 |
ENST00000366924.2 ENST00000308192.9 ENST00000418964.1 |
PLG |
plasminogen |
| chr7_-_107443652 | 0.14 |
ENST00000340010.5 ENST00000422236.2 ENST00000453332.1 |
SLC26A3 |
solute carrier family 26 (anion exchanger), member 3 |
| chrX_-_65259900 | 0.14 |
ENST00000412866.2 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr7_-_7782204 | 0.14 |
ENST00000418534.2 |
AC007161.5 |
AC007161.5 |
| chr14_+_93651358 | 0.13 |
ENST00000415050.2 |
TMEM251 |
transmembrane protein 251 |
| chr17_+_27895609 | 0.13 |
ENST00000581411.2 ENST00000301057.7 |
TP53I13 |
tumor protein p53 inducible protein 13 |
| chr3_-_64673668 | 0.13 |
ENST00000498707.1 |
ADAMTS9 |
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr11_-_62476965 | 0.13 |
ENST00000405837.1 ENST00000531524.1 |
BSCL2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr1_+_225600404 | 0.13 |
ENST00000366845.2 |
AC092811.1 |
AC092811.1 |
| chr2_+_67624430 | 0.13 |
ENST00000272342.5 |
ETAA1 |
Ewing tumor-associated antigen 1 |
| chr17_-_73975444 | 0.13 |
ENST00000293217.5 ENST00000537812.1 |
ACOX1 |
acyl-CoA oxidase 1, palmitoyl |
| chr17_+_56232494 | 0.13 |
ENST00000268912.5 |
OR4D1 |
olfactory receptor, family 4, subfamily D, member 1 |
| chrX_-_65253506 | 0.13 |
ENST00000427538.1 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr9_-_91793675 | 0.13 |
ENST00000375835.4 ENST00000375830.1 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr2_-_40657397 | 0.13 |
ENST00000408028.2 ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr12_+_80603233 | 0.13 |
ENST00000547103.1 ENST00000458043.2 |
OTOGL |
otogelin-like |
| chr11_-_62783276 | 0.13 |
ENST00000535878.1 ENST00000545207.1 |
SLC22A8 |
solute carrier family 22 (organic anion transporter), member 8 |
| chr12_-_82153087 | 0.12 |
ENST00000547623.1 ENST00000549396.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr22_+_25465786 | 0.12 |
ENST00000401395.1 |
KIAA1671 |
KIAA1671 |
| chr3_-_54962100 | 0.12 |
ENST00000273286.5 |
LRTM1 |
leucine-rich repeats and transmembrane domains 1 |
| chr15_+_31196053 | 0.11 |
ENST00000561594.1 ENST00000362065.4 |
FAN1 |
FANCD2/FANCI-associated nuclease 1 |
| chr14_+_22931924 | 0.11 |
ENST00000390477.2 |
TRDC |
T cell receptor delta constant |
| chr15_-_43559055 | 0.11 |
ENST00000220420.5 ENST00000349114.4 |
TGM5 |
transglutaminase 5 |
| chr2_-_207024233 | 0.11 |
ENST00000423725.1 ENST00000233190.6 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr7_-_107880508 | 0.11 |
ENST00000425651.2 |
NRCAM |
neuronal cell adhesion molecule |
| chr11_+_124481361 | 0.11 |
ENST00000284288.2 |
PANX3 |
pannexin 3 |
| chr3_+_138340049 | 0.11 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr2_+_74685527 | 0.11 |
ENST00000393972.3 ENST00000409737.1 ENST00000428943.1 |
WBP1 |
WW domain binding protein 1 |
| chr4_-_13546632 | 0.11 |
ENST00000382438.5 |
NKX3-2 |
NK3 homeobox 2 |
| chr1_-_154155595 | 0.11 |
ENST00000328159.4 ENST00000368531.2 ENST00000323144.7 ENST00000368533.3 ENST00000341372.3 |
TPM3 |
tropomyosin 3 |
| chr4_-_122085469 | 0.10 |
ENST00000057513.3 |
TNIP3 |
TNFAIP3 interacting protein 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 1.0 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.3 | 0.9 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.3 | 0.8 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.3 | 2.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.3 | 1.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.3 | 0.3 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.2 | 2.2 | GO:0001554 | luteolysis(GO:0001554) |
| 0.2 | 0.6 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.2 | 0.8 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 1.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.2 | 1.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.6 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 1.1 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 1.5 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 1.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.3 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.1 | 1.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 1.4 | GO:0048843 | positive regulation of axon extension involved in axon guidance(GO:0048842) negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.1 | 2.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.3 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 1.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 1.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 1.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 4.2 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 0.7 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.3 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.2 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 1.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 1.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 1.4 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.7 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.4 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 1.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.0 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.3 | GO:0008595 | blastoderm segmentation(GO:0007350) tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.5 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.3 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.4 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.3 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.9 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 2.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 1.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.3 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 1.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.7 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 2.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 1.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 1.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 2.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 2.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 1.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 1.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.2 | 0.9 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.5 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.1 | 1.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.3 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 1.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.3 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 7.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 3.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 1.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 1.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.6 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.8 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 4.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 2.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 2.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.9 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |