Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for NR3C1
Z-value: 0.28
Transcription factors associated with NR3C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR3C1
|
ENSG00000113580.10 | NR3C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR3C1 | hg19_v2_chr5_-_142783694_142783842, hg19_v2_chr5_-_142814241_142814278 | -0.32 | 4.3e-01 | Click! |
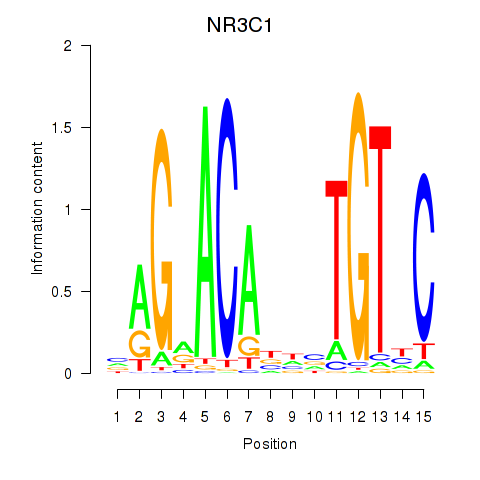
Activity profile of NR3C1 motif
Sorted Z-values of NR3C1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR3C1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_47352477 | 0.90 |
ENST00000593412.1 |
PRED62 |
Uncharacterized protein |
| chr3_-_182833863 | 0.83 |
ENST00000492597.1 |
MCCC1 |
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr18_-_500692 | 0.73 |
ENST00000400256.3 |
COLEC12 |
collectin sub-family member 12 |
| chr10_+_70847852 | 0.62 |
ENST00000242465.3 |
SRGN |
serglycin |
| chrX_-_119249819 | 0.59 |
ENST00000217999.2 |
RHOXF1 |
Rhox homeobox family, member 1 |
| chr22_-_17073700 | 0.59 |
ENST00000359963.3 |
CCT8L2 |
chaperonin containing TCP1, subunit 8 (theta)-like 2 |
| chr19_+_35630926 | 0.58 |
ENST00000588081.1 ENST00000589121.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr3_+_152017924 | 0.56 |
ENST00000465907.2 ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr4_-_70626314 | 0.53 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr21_-_46340807 | 0.50 |
ENST00000397846.3 ENST00000524251.1 ENST00000522688.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr8_+_91013676 | 0.49 |
ENST00000519410.1 ENST00000522161.1 ENST00000517761.1 ENST00000520227.1 |
DECR1 |
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr7_-_130353553 | 0.49 |
ENST00000330992.7 ENST00000445977.2 |
COPG2 |
coatomer protein complex, subunit gamma 2 |
| chr15_-_22448819 | 0.45 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr16_+_56716336 | 0.45 |
ENST00000394485.4 ENST00000562939.1 |
MT1X |
metallothionein 1X |
| chr11_+_117070904 | 0.45 |
ENST00000529792.1 |
TAGLN |
transgelin |
| chr3_-_119278376 | 0.45 |
ENST00000478182.1 |
CD80 |
CD80 molecule |
| chr7_+_76054224 | 0.45 |
ENST00000394857.3 |
ZP3 |
zona pellucida glycoprotein 3 (sperm receptor) |
| chr8_+_91013577 | 0.44 |
ENST00000220764.2 |
DECR1 |
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr1_-_183622442 | 0.44 |
ENST00000308641.4 |
APOBEC4 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative) |
| chr8_+_97597148 | 0.43 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr2_-_105372203 | 0.43 |
ENST00000424321.1 |
AC068057.2 |
long intergenic non-protein coding RNA 1114 |
| chr11_-_105948040 | 0.42 |
ENST00000534815.1 |
KBTBD3 |
kelch repeat and BTB (POZ) domain containing 3 |
| chr16_+_4674787 | 0.40 |
ENST00000262370.7 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr17_+_4618734 | 0.39 |
ENST00000571206.1 |
ARRB2 |
arrestin, beta 2 |
| chr16_+_56666563 | 0.39 |
ENST00000570233.1 |
MT1M |
metallothionein 1M |
| chr17_+_73975292 | 0.39 |
ENST00000397640.1 ENST00000416485.1 ENST00000588202.1 ENST00000590676.1 ENST00000586891.1 |
TEN1 |
TEN1 CST complex subunit |
| chr4_+_55095264 | 0.39 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr19_-_10445399 | 0.38 |
ENST00000592945.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr22_+_25003568 | 0.38 |
ENST00000447416.1 |
GGT1 |
gamma-glutamyltransferase 1 |
| chr11_-_105948129 | 0.38 |
ENST00000526793.1 |
KBTBD3 |
kelch repeat and BTB (POZ) domain containing 3 |
| chr2_+_233925064 | 0.37 |
ENST00000359570.5 ENST00000538935.1 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
| chr3_-_190167571 | 0.37 |
ENST00000354905.2 |
TMEM207 |
transmembrane protein 207 |
| chr10_-_27529486 | 0.37 |
ENST00000375888.1 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr11_-_63933504 | 0.37 |
ENST00000255681.6 |
MACROD1 |
MACRO domain containing 1 |
| chr16_-_88770019 | 0.37 |
ENST00000541206.2 |
RNF166 |
ring finger protein 166 |
| chr1_-_27816641 | 0.37 |
ENST00000430629.2 |
WASF2 |
WAS protein family, member 2 |
| chr13_+_102142296 | 0.37 |
ENST00000376162.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr6_+_26045603 | 0.36 |
ENST00000540144.1 |
HIST1H3C |
histone cluster 1, H3c |
| chr12_-_110486348 | 0.36 |
ENST00000547573.1 ENST00000546651.2 ENST00000551185.2 |
C12orf76 |
chromosome 12 open reading frame 76 |
| chr1_+_151735431 | 0.35 |
ENST00000321531.5 ENST00000315067.8 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr17_-_19290117 | 0.35 |
ENST00000497081.2 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr10_-_28571015 | 0.34 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_-_27816556 | 0.33 |
ENST00000536657.1 |
WASF2 |
WAS protein family, member 2 |
| chr12_+_53443963 | 0.32 |
ENST00000546602.1 ENST00000552570.1 ENST00000549700.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr1_+_70876926 | 0.32 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr8_-_100025238 | 0.32 |
ENST00000521696.1 |
RP11-410L14.2 |
RP11-410L14.2 |
| chr19_-_10450328 | 0.32 |
ENST00000160262.5 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr15_+_31658349 | 0.32 |
ENST00000558844.1 |
KLF13 |
Kruppel-like factor 13 |
| chr12_-_58329819 | 0.31 |
ENST00000551421.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr11_-_114271139 | 0.31 |
ENST00000325636.4 |
C11orf71 |
chromosome 11 open reading frame 71 |
| chr17_-_67138015 | 0.31 |
ENST00000284425.2 ENST00000590645.1 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr17_-_19290483 | 0.31 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr1_+_28844778 | 0.30 |
ENST00000411533.1 |
RCC1 |
regulator of chromosome condensation 1 |
| chr19_-_10450287 | 0.30 |
ENST00000589261.1 ENST00000590569.1 ENST00000589580.1 ENST00000589249.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr1_-_85870177 | 0.30 |
ENST00000542148.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr11_+_12115543 | 0.30 |
ENST00000537344.1 ENST00000532179.1 ENST00000526065.1 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr10_-_61469837 | 0.30 |
ENST00000395348.3 |
SLC16A9 |
solute carrier family 16, member 9 |
| chr19_-_58864848 | 0.29 |
ENST00000263100.3 |
A1BG |
alpha-1-B glycoprotein |
| chr16_-_33647696 | 0.29 |
ENST00000558425.1 ENST00000569103.2 |
RP11-812E19.9 |
Uncharacterized protein |
| chr4_-_681114 | 0.29 |
ENST00000503156.1 |
MFSD7 |
major facilitator superfamily domain containing 7 |
| chr17_-_73761222 | 0.29 |
ENST00000437911.1 ENST00000225614.2 |
GALK1 |
galactokinase 1 |
| chr1_-_85930246 | 0.29 |
ENST00000426972.3 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr21_-_34185944 | 0.29 |
ENST00000479548.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr5_-_172198190 | 0.29 |
ENST00000239223.3 |
DUSP1 |
dual specificity phosphatase 1 |
| chr2_-_158300556 | 0.29 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr12_-_47219733 | 0.29 |
ENST00000547477.1 ENST00000447411.1 ENST00000266579.4 |
SLC38A4 |
solute carrier family 38, member 4 |
| chr2_-_79315112 | 0.29 |
ENST00000305089.3 |
REG1B |
regenerating islet-derived 1 beta |
| chr10_-_28623368 | 0.28 |
ENST00000441595.2 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_-_182360498 | 0.28 |
ENST00000417584.2 |
GLUL |
glutamate-ammonia ligase |
| chr21_-_46221684 | 0.28 |
ENST00000330942.5 |
UBE2G2 |
ubiquitin-conjugating enzyme E2G 2 |
| chr16_+_1578674 | 0.28 |
ENST00000253934.5 |
TMEM204 |
transmembrane protein 204 |
| chr16_+_4674814 | 0.28 |
ENST00000415496.1 ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr6_-_37467628 | 0.27 |
ENST00000373408.3 |
CCDC167 |
coiled-coil domain containing 167 |
| chr17_+_66245341 | 0.27 |
ENST00000577985.1 |
AMZ2 |
archaelysin family metallopeptidase 2 |
| chr11_+_124609823 | 0.27 |
ENST00000412681.2 |
NRGN |
neurogranin (protein kinase C substrate, RC3) |
| chr1_-_182360918 | 0.27 |
ENST00000339526.4 |
GLUL |
glutamate-ammonia ligase |
| chr18_-_19994830 | 0.27 |
ENST00000525417.1 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr14_-_61124977 | 0.27 |
ENST00000554986.1 |
SIX1 |
SIX homeobox 1 |
| chr19_+_35630628 | 0.27 |
ENST00000588715.1 ENST00000588607.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr1_+_207262170 | 0.27 |
ENST00000367078.3 |
C4BPB |
complement component 4 binding protein, beta |
| chr4_-_152149033 | 0.26 |
ENST00000514152.1 |
SH3D19 |
SH3 domain containing 19 |
| chr1_+_28844648 | 0.26 |
ENST00000373832.1 ENST00000373831.3 |
RCC1 |
regulator of chromosome condensation 1 |
| chr3_+_12330560 | 0.26 |
ENST00000397026.2 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr3_+_9944303 | 0.26 |
ENST00000421412.1 ENST00000295980.3 |
IL17RE |
interleukin 17 receptor E |
| chr5_+_67588391 | 0.26 |
ENST00000523872.1 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr16_-_67260691 | 0.26 |
ENST00000447579.1 ENST00000393992.1 ENST00000424285.1 |
LRRC29 |
leucine rich repeat containing 29 |
| chr2_+_234637754 | 0.26 |
ENST00000482026.1 ENST00000609767.1 |
UGT1A3 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr17_+_75401152 | 0.25 |
ENST00000585930.1 |
SEPT9 |
septin 9 |
| chr6_+_18387570 | 0.25 |
ENST00000259939.3 |
RNF144B |
ring finger protein 144B |
| chr6_+_132873832 | 0.25 |
ENST00000275200.1 |
TAAR8 |
trace amine associated receptor 8 |
| chr1_+_207262578 | 0.25 |
ENST00000243611.5 ENST00000367076.3 |
C4BPB |
complement component 4 binding protein, beta |
| chr7_-_76255444 | 0.25 |
ENST00000454397.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr14_+_105935396 | 0.25 |
ENST00000494981.1 |
MTA1 |
metastasis associated 1 |
| chr3_-_58563094 | 0.25 |
ENST00000464064.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr15_+_33010175 | 0.24 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr1_+_207262627 | 0.24 |
ENST00000391923.1 |
C4BPB |
complement component 4 binding protein, beta |
| chr19_-_2456922 | 0.24 |
ENST00000582871.1 ENST00000325327.3 |
LMNB2 |
lamin B2 |
| chr7_-_19157248 | 0.24 |
ENST00000242261.5 |
TWIST1 |
twist family bHLH transcription factor 1 |
| chr5_-_141392538 | 0.24 |
ENST00000503794.1 ENST00000510194.1 ENST00000504424.1 ENST00000513454.1 ENST00000458112.2 ENST00000542860.1 ENST00000503229.1 ENST00000500692.2 ENST00000311337.6 ENST00000504139.1 ENST00000505689.1 |
GNPDA1 |
glucosamine-6-phosphate deaminase 1 |
| chr14_-_23426270 | 0.24 |
ENST00000557591.1 ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr20_+_138089 | 0.24 |
ENST00000382388.3 |
DEFB127 |
defensin, beta 127 |
| chr15_+_97326659 | 0.24 |
ENST00000558553.1 |
SPATA8 |
spermatogenesis associated 8 |
| chr3_-_121379739 | 0.23 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr7_+_150688083 | 0.23 |
ENST00000297494.3 |
NOS3 |
nitric oxide synthase 3 (endothelial cell) |
| chr16_+_618837 | 0.23 |
ENST00000409439.2 |
PIGQ |
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr19_-_42916499 | 0.23 |
ENST00000601189.1 ENST00000599211.1 |
LIPE |
lipase, hormone-sensitive |
| chr1_-_38218577 | 0.23 |
ENST00000540011.1 |
EPHA10 |
EPH receptor A10 |
| chr6_+_33048222 | 0.23 |
ENST00000428835.1 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
| chr16_-_11370330 | 0.23 |
ENST00000241808.4 ENST00000435245.2 |
PRM2 |
protamine 2 |
| chr17_-_55911970 | 0.23 |
ENST00000581805.1 ENST00000580960.1 |
RP11-60A24.3 |
RP11-60A24.3 |
| chr8_-_30585439 | 0.23 |
ENST00000221130.5 |
GSR |
glutathione reductase |
| chr16_+_67207838 | 0.23 |
ENST00000566871.1 ENST00000268605.7 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr1_+_207262540 | 0.22 |
ENST00000452902.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr2_-_191878162 | 0.22 |
ENST00000540176.1 |
STAT1 |
signal transducer and activator of transcription 1, 91kDa |
| chr3_-_75834722 | 0.22 |
ENST00000471541.2 |
ZNF717 |
zinc finger protein 717 |
| chr10_-_76868931 | 0.22 |
ENST00000372700.3 ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13 |
dual specificity phosphatase 13 |
| chr18_-_11670159 | 0.22 |
ENST00000561598.1 |
RP11-677O4.2 |
RP11-677O4.2 |
| chr15_+_55700741 | 0.22 |
ENST00000569691.1 |
C15orf65 |
chromosome 15 open reading frame 65 |
| chr2_-_69180012 | 0.22 |
ENST00000481498.1 |
GKN2 |
gastrokine 2 |
| chr20_-_5426332 | 0.22 |
ENST00000420529.1 |
LINC00658 |
long intergenic non-protein coding RNA 658 |
| chr17_+_48911942 | 0.22 |
ENST00000426127.1 |
WFIKKN2 |
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr22_-_27456361 | 0.22 |
ENST00000453934.1 |
CTA-992D9.6 |
CTA-992D9.6 |
| chr16_+_67207872 | 0.22 |
ENST00000563258.1 ENST00000568146.1 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr14_-_106845789 | 0.22 |
ENST00000390617.2 |
IGHV3-35 |
immunoglobulin heavy variable 3-35 (non-functional) |
| chr14_-_23426337 | 0.22 |
ENST00000342454.8 ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr2_+_10560147 | 0.21 |
ENST00000422133.1 |
HPCAL1 |
hippocalcin-like 1 |
| chr14_-_53258314 | 0.21 |
ENST00000216410.3 ENST00000557604.1 |
GNPNAT1 |
glucosamine-phosphate N-acetyltransferase 1 |
| chr14_-_100841670 | 0.21 |
ENST00000557297.1 ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS |
tryptophanyl-tRNA synthetase |
| chr9_+_86238016 | 0.21 |
ENST00000530832.1 ENST00000405990.3 ENST00000376417.4 ENST00000376419.4 |
IDNK |
idnK, gluconokinase homolog (E. coli) |
| chr17_+_7461613 | 0.21 |
ENST00000438470.1 ENST00000436057.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr14_-_23426322 | 0.21 |
ENST00000555367.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr14_-_106725723 | 0.21 |
ENST00000390609.2 |
IGHV3-23 |
immunoglobulin heavy variable 3-23 |
| chr8_+_26247878 | 0.21 |
ENST00000518611.1 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr17_-_45056606 | 0.21 |
ENST00000322329.3 |
RPRML |
reprimo-like |
| chr7_+_117864708 | 0.21 |
ENST00000357099.4 ENST00000265224.4 ENST00000486422.1 ENST00000417525.1 |
ANKRD7 |
ankyrin repeat domain 7 |
| chr19_+_12862604 | 0.21 |
ENST00000553030.1 |
BEST2 |
bestrophin 2 |
| chr8_-_121457332 | 0.21 |
ENST00000518918.1 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr19_+_35630344 | 0.21 |
ENST00000455515.2 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr17_+_40704938 | 0.21 |
ENST00000225929.5 |
HSD17B1 |
hydroxysteroid (17-beta) dehydrogenase 1 |
| chr11_-_34533257 | 0.21 |
ENST00000312319.2 |
ELF5 |
E74-like factor 5 (ets domain transcription factor) |
| chr10_-_27529716 | 0.21 |
ENST00000375897.3 ENST00000396271.3 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr5_-_76383133 | 0.20 |
ENST00000255198.2 |
ZBED3 |
zinc finger, BED-type containing 3 |
| chr4_-_70626430 | 0.20 |
ENST00000310613.3 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr1_+_158969752 | 0.20 |
ENST00000566111.1 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr8_-_27469196 | 0.20 |
ENST00000546343.1 ENST00000560566.1 |
CLU |
clusterin |
| chr16_+_33020496 | 0.20 |
ENST00000565407.2 |
IGHV3OR16-8 |
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr10_+_134351319 | 0.20 |
ENST00000368594.3 ENST00000368593.3 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
| chr2_-_42991257 | 0.20 |
ENST00000378661.2 |
OXER1 |
oxoeicosanoid (OXE) receptor 1 |
| chr18_+_657578 | 0.19 |
ENST00000323274.10 |
TYMS |
thymidylate synthetase |
| chr17_-_15496722 | 0.19 |
ENST00000472534.1 |
CDRT1 |
CMT1A duplicated region transcript 1 |
| chr11_-_2924970 | 0.19 |
ENST00000533594.1 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr11_-_2924720 | 0.19 |
ENST00000455942.2 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr12_-_50290839 | 0.19 |
ENST00000552863.1 |
FAIM2 |
Fas apoptotic inhibitory molecule 2 |
| chr19_+_5823813 | 0.19 |
ENST00000303212.2 |
NRTN |
neurturin |
| chr14_+_60715928 | 0.19 |
ENST00000395076.4 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr12_+_53443680 | 0.19 |
ENST00000314250.6 ENST00000451358.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr1_-_36863481 | 0.19 |
ENST00000315732.2 |
LSM10 |
LSM10, U7 small nuclear RNA associated |
| chr14_+_105190514 | 0.19 |
ENST00000330877.2 |
ADSSL1 |
adenylosuccinate synthase like 1 |
| chr22_-_24322660 | 0.19 |
ENST00000404092.1 |
DDT |
D-dopachrome tautomerase |
| chr13_+_103459704 | 0.19 |
ENST00000602836.1 |
BIVM-ERCC5 |
BIVM-ERCC5 readthrough |
| chr16_+_56659687 | 0.19 |
ENST00000568293.1 ENST00000330439.6 |
MT1E |
metallothionein 1E |
| chrY_+_16168097 | 0.19 |
ENST00000250823.4 |
VCY1B |
variable charge, Y-linked 1B |
| chr18_-_5296138 | 0.19 |
ENST00000400143.3 |
ZBTB14 |
zinc finger and BTB domain containing 14 |
| chr22_-_19435209 | 0.19 |
ENST00000546308.1 ENST00000541063.1 ENST00000399568.1 ENST00000333059.5 |
HIRA C22orf39 |
histone cell cycle regulator chromosome 22 open reading frame 39 |
| chr15_-_48937982 | 0.19 |
ENST00000316623.5 |
FBN1 |
fibrillin 1 |
| chr3_-_142720267 | 0.19 |
ENST00000597953.1 |
RP11-91G21.1 |
RP11-91G21.1 |
| chr14_-_24685246 | 0.19 |
ENST00000396833.2 ENST00000288087.7 |
MDP1 |
magnesium-dependent phosphatase 1 |
| chr9_+_127615733 | 0.19 |
ENST00000373574.1 |
WDR38 |
WD repeat domain 38 |
| chr22_-_37172111 | 0.19 |
ENST00000417951.2 ENST00000430701.1 ENST00000433985.2 |
IFT27 |
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr4_-_89744314 | 0.18 |
ENST00000508369.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr6_-_31648150 | 0.18 |
ENST00000375858.3 ENST00000383237.4 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr8_-_101661887 | 0.18 |
ENST00000311812.2 |
SNX31 |
sorting nexin 31 |
| chr14_+_74353320 | 0.18 |
ENST00000540593.1 ENST00000555730.1 |
ZNF410 |
zinc finger protein 410 |
| chr1_+_207262881 | 0.18 |
ENST00000451804.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr22_-_37172191 | 0.18 |
ENST00000340630.5 |
IFT27 |
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr14_+_88851874 | 0.18 |
ENST00000393545.4 ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7 |
spermatogenesis associated 7 |
| chr11_+_65266507 | 0.18 |
ENST00000544868.1 |
MALAT1 |
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr21_-_31744557 | 0.18 |
ENST00000399889.2 |
KRTAP13-2 |
keratin associated protein 13-2 |
| chr2_+_114163945 | 0.18 |
ENST00000453673.3 |
IGKV1OR2-108 |
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr13_-_38172863 | 0.18 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr1_+_15632231 | 0.18 |
ENST00000375997.4 ENST00000524761.1 ENST00000375995.3 ENST00000401090.2 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr1_-_51425902 | 0.18 |
ENST00000396153.2 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
| chr11_-_102826434 | 0.18 |
ENST00000340273.4 ENST00000260302.3 |
MMP13 |
matrix metallopeptidase 13 (collagenase 3) |
| chr17_-_8027402 | 0.18 |
ENST00000541682.2 ENST00000317814.4 ENST00000577735.1 |
HES7 |
hes family bHLH transcription factor 7 |
| chr17_+_7905912 | 0.18 |
ENST00000254854.4 |
GUCY2D |
guanylate cyclase 2D, membrane (retina-specific) |
| chr9_+_131683174 | 0.18 |
ENST00000372592.3 ENST00000428610.1 |
PHYHD1 |
phytanoyl-CoA dioxygenase domain containing 1 |
| chr2_-_89278535 | 0.17 |
ENST00000390247.2 |
IGKV3-7 |
immunoglobulin kappa variable 3-7 (non-functional) |
| chr3_-_119278446 | 0.17 |
ENST00000264246.3 |
CD80 |
CD80 molecule |
| chr11_-_65686586 | 0.17 |
ENST00000438576.2 |
C11orf68 |
chromosome 11 open reading frame 68 |
| chr6_+_88032299 | 0.17 |
ENST00000608353.1 ENST00000392863.1 ENST00000229570.5 ENST00000608525.1 ENST00000608868.1 |
SMIM8 |
small integral membrane protein 8 |
| chr15_+_42787815 | 0.17 |
ENST00000564153.1 ENST00000349777.1 ENST00000567094.1 ENST00000566327.1 ENST00000568859.1 |
SNAP23 |
synaptosomal-associated protein, 23kDa |
| chr4_-_89744365 | 0.17 |
ENST00000513837.1 ENST00000503556.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr2_+_27719697 | 0.17 |
ENST00000264717.2 ENST00000424318.2 |
GCKR |
glucokinase (hexokinase 4) regulator |
| chr7_+_150688166 | 0.17 |
ENST00000461406.1 |
NOS3 |
nitric oxide synthase 3 (endothelial cell) |
| chr4_-_107957454 | 0.17 |
ENST00000285311.3 |
DKK2 |
dickkopf WNT signaling pathway inhibitor 2 |
| chr18_+_56530136 | 0.17 |
ENST00000591083.1 |
ZNF532 |
zinc finger protein 532 |
| chr12_-_53074182 | 0.17 |
ENST00000252244.3 |
KRT1 |
keratin 1 |
| chr9_+_132962843 | 0.17 |
ENST00000458469.1 |
NCS1 |
neuronal calcium sensor 1 |
| chr4_-_140222358 | 0.17 |
ENST00000505036.1 ENST00000544855.1 ENST00000539002.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr16_+_12058961 | 0.17 |
ENST00000053243.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 0.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 1.1 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.4 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.1 | 0.4 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 0.4 | GO:0014806 | negative regulation of muscle hyperplasia(GO:0014740) smooth muscle hyperplasia(GO:0014806) |
| 0.1 | 1.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.1 | 0.4 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.4 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.1 | 0.4 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.4 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.6 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 0.6 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.8 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.3 | GO:0019860 | transformation of host cell by virus(GO:0019087) uracil metabolic process(GO:0019860) |
| 0.1 | 0.2 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.3 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.2 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.4 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.9 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.3 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.2 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.1 | 0.4 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.3 | GO:2000230 | response to metformin(GO:1901558) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.2 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.1 | 0.5 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.2 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.1 | 0.5 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.1 | 0.2 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.2 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.1 | 0.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.7 | GO:0071493 | elastic fiber assembly(GO:0048251) cellular response to UV-B(GO:0071493) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.5 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:1902356 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.6 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.7 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.4 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:0032672 | regulation of interleukin-3 production(GO:0032672) |
| 0.0 | 0.2 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.0 | 0.7 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.3 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.2 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.2 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.3 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.3 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.3 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0042418 | epinephrine metabolic process(GO:0042414) epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.7 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.1 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.1 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.0 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.6 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0048936 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.1 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:2000322 | regulation of glucocorticoid receptor signaling pathway(GO:2000322) |
| 0.0 | 0.0 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0032916 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.3 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.0 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.0 | 0.0 | GO:1902310 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.5 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.2 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.2 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.1 | GO:1903147 | negative regulation of macromitophagy(GO:1901525) negative regulation of mitophagy(GO:1903147) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.4 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 1.2 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0097242 | beta-amyloid clearance(GO:0097242) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0033133 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.0 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 2.0 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.1 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.1 | GO:0060180 | female mating behavior(GO:0060180) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 0.4 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.1 | 0.2 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 0.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.5 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0071749 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.0 | 0.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.6 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.4 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.0 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 1.2 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.2 | 0.6 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 0.8 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.6 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.4 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 1.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.2 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.2 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 0.2 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.5 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.2 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.5 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.0 | 0.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 1.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0004979 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.0 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 1.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.8 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0004609 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.8 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.0 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.0 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |