Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for NR4A1
Z-value: 0.78
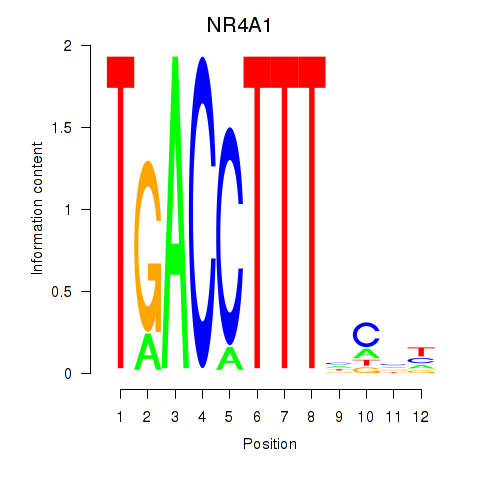
Transcription factors associated with NR4A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A1
|
ENSG00000123358.15 | NR4A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A1 | hg19_v2_chr12_+_52445191_52445243 | -0.25 | 5.5e-01 | Click! |
Activity profile of NR4A1 motif
Sorted Z-values of NR4A1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_120685608 | 0.89 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr20_+_12989596 | 0.31 |
ENST00000434210.1 ENST00000399002.2 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
| chr6_-_152489484 | 0.30 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr1_-_161102367 | 0.27 |
ENST00000464113.1 |
DEDD |
death effector domain containing |
| chr10_-_33625154 | 0.26 |
ENST00000265371.4 |
NRP1 |
neuropilin 1 |
| chr19_+_14138960 | 0.25 |
ENST00000431365.2 ENST00000585987.1 |
RLN3 |
relaxin 3 |
| chr15_+_75335604 | 0.25 |
ENST00000563393.1 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
| chr3_+_29322437 | 0.25 |
ENST00000434693.2 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
| chr14_+_29236269 | 0.24 |
ENST00000313071.4 |
FOXG1 |
forkhead box G1 |
| chr12_+_32655048 | 0.22 |
ENST00000427716.2 ENST00000266482.3 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr4_-_159080806 | 0.21 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr16_+_53241854 | 0.21 |
ENST00000565803.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr20_+_37590942 | 0.21 |
ENST00000373325.2 ENST00000252011.3 ENST00000373323.4 |
DHX35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr4_+_71108300 | 0.20 |
ENST00000304954.3 |
CSN3 |
casein kappa |
| chr20_-_42816206 | 0.20 |
ENST00000372980.3 |
JPH2 |
junctophilin 2 |
| chr7_-_105517021 | 0.20 |
ENST00000318724.4 ENST00000419735.3 |
ATXN7L1 |
ataxin 7-like 1 |
| chr6_-_152958521 | 0.19 |
ENST00000367255.5 ENST00000265368.4 ENST00000448038.1 ENST00000341594.5 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr20_+_17680587 | 0.19 |
ENST00000427254.1 ENST00000377805.3 |
BANF2 |
barrier to autointegration factor 2 |
| chr12_+_21679220 | 0.18 |
ENST00000256969.2 |
C12orf39 |
chromosome 12 open reading frame 39 |
| chr20_-_42815733 | 0.17 |
ENST00000342272.3 |
JPH2 |
junctophilin 2 |
| chr4_+_100495864 | 0.17 |
ENST00000265517.5 ENST00000422897.2 |
MTTP |
microsomal triglyceride transfer protein |
| chr2_+_162016827 | 0.16 |
ENST00000429217.1 ENST00000406287.1 ENST00000402568.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr6_+_69942298 | 0.16 |
ENST00000238918.8 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
| chr10_-_61666267 | 0.16 |
ENST00000263102.6 |
CCDC6 |
coiled-coil domain containing 6 |
| chr20_-_44485835 | 0.15 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr19_+_54926601 | 0.15 |
ENST00000301194.4 |
TTYH1 |
tweety family member 1 |
| chr19_+_54926621 | 0.15 |
ENST00000376530.3 ENST00000445095.1 ENST00000391739.3 ENST00000376531.3 |
TTYH1 |
tweety family member 1 |
| chr1_+_61869748 | 0.15 |
ENST00000357977.5 |
NFIA |
nuclear factor I/A |
| chr20_+_11898507 | 0.15 |
ENST00000378226.2 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr2_+_178257372 | 0.15 |
ENST00000264167.4 ENST00000409888.1 |
AGPS |
alkylglycerone phosphate synthase |
| chr1_-_161102421 | 0.14 |
ENST00000490843.2 ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD |
death effector domain containing |
| chr12_+_96252706 | 0.14 |
ENST00000266735.5 ENST00000553192.1 ENST00000552085.1 |
SNRPF |
small nuclear ribonucleoprotein polypeptide F |
| chr11_+_116700600 | 0.14 |
ENST00000227667.3 |
APOC3 |
apolipoprotein C-III |
| chr7_-_135612198 | 0.13 |
ENST00000589735.1 |
LUZP6 |
leucine zipper protein 6 |
| chr11_+_116700614 | 0.13 |
ENST00000375345.1 |
APOC3 |
apolipoprotein C-III |
| chr4_-_89744314 | 0.13 |
ENST00000508369.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr2_+_162016804 | 0.13 |
ENST00000392749.2 ENST00000440506.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr10_+_35484793 | 0.13 |
ENST00000488741.1 ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM |
cAMP responsive element modulator |
| chr7_-_72936608 | 0.13 |
ENST00000404251.1 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
| chr4_-_89744365 | 0.12 |
ENST00000513837.1 ENST00000503556.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr7_-_6048650 | 0.12 |
ENST00000382321.4 ENST00000406569.3 |
PMS2 |
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr2_+_162016916 | 0.12 |
ENST00000405852.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr17_-_2169425 | 0.12 |
ENST00000570606.1 ENST00000354901.4 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
| chr1_+_104293028 | 0.11 |
ENST00000370079.3 |
AMY1C |
amylase, alpha 1C (salivary) |
| chr16_+_81272287 | 0.11 |
ENST00000425577.2 ENST00000564552.1 |
BCMO1 |
beta-carotene 15,15'-monooxygenase 1 |
| chr7_-_72936531 | 0.11 |
ENST00000339594.4 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
| chr6_+_64346386 | 0.11 |
ENST00000509330.1 |
PHF3 |
PHD finger protein 3 |
| chr6_-_43595039 | 0.11 |
ENST00000307114.7 |
GTPBP2 |
GTP binding protein 2 |
| chr6_+_44126545 | 0.11 |
ENST00000532171.1 ENST00000398776.1 ENST00000542245.1 |
CAPN11 |
calpain 11 |
| chr19_+_17416609 | 0.11 |
ENST00000602206.1 |
MRPL34 |
mitochondrial ribosomal protein L34 |
| chr17_-_7123021 | 0.11 |
ENST00000399510.2 |
DLG4 |
discs, large homolog 4 (Drosophila) |
| chr6_+_36973406 | 0.11 |
ENST00000274963.8 |
FGD2 |
FYVE, RhoGEF and PH domain containing 2 |
| chr1_+_165600436 | 0.10 |
ENST00000367888.4 ENST00000367885.1 ENST00000367884.2 |
MGST3 |
microsomal glutathione S-transferase 3 |
| chr12_+_109577202 | 0.10 |
ENST00000377848.3 ENST00000377854.5 |
ACACB |
acetyl-CoA carboxylase beta |
| chr17_+_4854375 | 0.10 |
ENST00000521811.1 ENST00000519602.1 ENST00000323997.6 ENST00000522249.1 ENST00000519584.1 |
ENO3 |
enolase 3 (beta, muscle) |
| chr22_+_24891210 | 0.10 |
ENST00000382760.2 |
UPB1 |
ureidopropionase, beta |
| chr19_+_17416457 | 0.10 |
ENST00000252602.1 |
MRPL34 |
mitochondrial ribosomal protein L34 |
| chr13_-_30881134 | 0.10 |
ENST00000380617.3 ENST00000441394.1 |
KATNAL1 |
katanin p60 subunit A-like 1 |
| chr1_+_40713573 | 0.09 |
ENST00000372766.3 |
TMCO2 |
transmembrane and coiled-coil domains 2 |
| chr7_-_6048702 | 0.09 |
ENST00000265849.7 |
PMS2 |
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr11_-_111794446 | 0.09 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr11_-_113577014 | 0.09 |
ENST00000544634.1 ENST00000539732.1 ENST00000538770.1 ENST00000536856.1 ENST00000544476.1 |
TMPRSS5 |
transmembrane protease, serine 5 |
| chr3_-_113464906 | 0.09 |
ENST00000477813.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr8_-_92053212 | 0.09 |
ENST00000285419.3 |
TMEM55A |
transmembrane protein 55A |
| chr4_+_159593271 | 0.09 |
ENST00000512251.1 ENST00000511912.1 |
ETFDH |
electron-transferring-flavoprotein dehydrogenase |
| chr19_+_6372444 | 0.08 |
ENST00000245812.3 |
ALKBH7 |
alkB, alkylation repair homolog 7 (E. coli) |
| chr17_+_7123125 | 0.08 |
ENST00000356839.5 ENST00000583312.1 ENST00000350303.5 |
ACADVL |
acyl-CoA dehydrogenase, very long chain |
| chr5_-_169626104 | 0.08 |
ENST00000520275.1 ENST00000506431.2 |
CTB-27N1.1 |
CTB-27N1.1 |
| chr11_-_113577052 | 0.08 |
ENST00000540540.1 ENST00000545579.1 ENST00000538955.1 ENST00000299882.5 |
TMPRSS5 |
transmembrane protease, serine 5 |
| chr8_+_99439214 | 0.08 |
ENST00000287042.4 |
KCNS2 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
| chr7_-_105516923 | 0.08 |
ENST00000478915.1 |
ATXN7L1 |
ataxin 7-like 1 |
| chr12_+_6644443 | 0.08 |
ENST00000396858.1 |
GAPDH |
glyceraldehyde-3-phosphate dehydrogenase |
| chr7_-_81399438 | 0.08 |
ENST00000222390.5 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr10_+_101542462 | 0.08 |
ENST00000370449.4 ENST00000370434.1 |
ABCC2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr16_+_691792 | 0.07 |
ENST00000307650.4 |
FAM195A |
family with sequence similarity 195, member A |
| chr2_+_234216454 | 0.07 |
ENST00000447536.1 ENST00000409110.1 |
SAG |
S-antigen; retina and pineal gland (arrestin) |
| chr3_-_48936272 | 0.07 |
ENST00000544097.1 ENST00000430379.1 ENST00000319017.4 |
SLC25A20 |
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr20_+_17680599 | 0.07 |
ENST00000246090.5 |
BANF2 |
barrier to autointegration factor 2 |
| chr4_-_57253587 | 0.07 |
ENST00000513376.1 ENST00000602986.1 ENST00000434343.2 ENST00000451613.1 ENST00000205214.6 ENST00000502617.1 |
AASDH |
aminoadipate-semialdehyde dehydrogenase |
| chr14_-_104387831 | 0.06 |
ENST00000557040.1 ENST00000414262.2 ENST00000555030.1 ENST00000554713.1 ENST00000553430.1 |
C14orf2 |
chromosome 14 open reading frame 2 |
| chr17_-_79283041 | 0.06 |
ENST00000332012.5 |
LINC00482 |
long intergenic non-protein coding RNA 482 |
| chr1_+_16062820 | 0.06 |
ENST00000294454.5 |
SLC25A34 |
solute carrier family 25, member 34 |
| chr12_+_54378923 | 0.06 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr19_-_41256207 | 0.06 |
ENST00000598485.2 ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54 |
chromosome 19 open reading frame 54 |
| chr13_-_33112823 | 0.06 |
ENST00000504114.1 |
N4BP2L2 |
NEDD4 binding protein 2-like 2 |
| chr4_-_89744457 | 0.06 |
ENST00000395002.2 |
FAM13A |
family with sequence similarity 13, member A |
| chr3_-_44519131 | 0.06 |
ENST00000425708.2 ENST00000396077.2 |
ZNF445 |
zinc finger protein 445 |
| chr17_-_7307358 | 0.06 |
ENST00000576017.1 ENST00000302422.3 ENST00000535512.1 |
TMEM256 TMEM256-PLSCR3 |
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr10_-_75634219 | 0.05 |
ENST00000305762.7 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
| chr14_-_104387888 | 0.05 |
ENST00000286953.3 |
C14orf2 |
chromosome 14 open reading frame 2 |
| chr7_-_81399355 | 0.05 |
ENST00000457544.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr14_-_35591433 | 0.05 |
ENST00000261475.5 ENST00000555644.1 |
PPP2R3C |
protein phosphatase 2, regulatory subunit B'', gamma |
| chr14_-_77495007 | 0.05 |
ENST00000238647.3 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
| chr17_+_4855053 | 0.05 |
ENST00000518175.1 |
ENO3 |
enolase 3 (beta, muscle) |
| chr12_+_58335360 | 0.05 |
ENST00000300145.3 |
XRCC6BP1 |
XRCC6 binding protein 1 |
| chr4_-_74088800 | 0.05 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr4_-_40632605 | 0.05 |
ENST00000514014.1 |
RBM47 |
RNA binding motif protein 47 |
| chr5_+_159895275 | 0.04 |
ENST00000517927.1 |
MIR146A |
microRNA 146a |
| chr10_-_74714533 | 0.04 |
ENST00000373032.3 |
PLA2G12B |
phospholipase A2, group XIIB |
| chr2_-_25194963 | 0.04 |
ENST00000264711.2 |
DNAJC27 |
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr3_+_113465866 | 0.04 |
ENST00000273398.3 ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A |
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr17_+_79670386 | 0.04 |
ENST00000333676.3 ENST00000571730.1 ENST00000541223.1 |
MRPL12 SLC25A10 SLC25A10 |
mitochondrial ribosomal protein L12 Mitochondrial dicarboxylate carrier; Uncharacterized protein; cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr14_-_24711470 | 0.04 |
ENST00000559969.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr3_+_138340049 | 0.04 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr18_-_60986613 | 0.04 |
ENST00000444484.1 |
BCL2 |
B-cell CLL/lymphoma 2 |
| chr6_+_31895254 | 0.04 |
ENST00000299367.5 ENST00000442278.2 |
C2 |
complement component 2 |
| chr14_+_22386325 | 0.04 |
ENST00000390439.2 |
TRAV13-2 |
T cell receptor alpha variable 13-2 |
| chr22_-_51016846 | 0.04 |
ENST00000312108.7 ENST00000395650.2 |
CPT1B |
carnitine palmitoyltransferase 1B (muscle) |
| chr2_+_228678550 | 0.03 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chr1_+_50569575 | 0.03 |
ENST00000371827.1 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr12_+_21525818 | 0.03 |
ENST00000240652.3 ENST00000542023.1 ENST00000537593.1 |
IAPP |
islet amyloid polypeptide |
| chr12_+_93963590 | 0.03 |
ENST00000340600.2 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr6_+_118869452 | 0.03 |
ENST00000357525.5 |
PLN |
phospholamban |
| chr6_-_33771757 | 0.03 |
ENST00000507738.1 ENST00000266003.5 ENST00000430124.2 |
MLN |
motilin |
| chr22_-_51017084 | 0.03 |
ENST00000360719.2 ENST00000457250.1 ENST00000440709.1 |
CPT1B |
carnitine palmitoyltransferase 1B (muscle) |
| chr6_+_31895480 | 0.03 |
ENST00000418949.2 ENST00000383177.3 ENST00000477310.1 |
C2 CFB |
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr3_+_101443476 | 0.03 |
ENST00000327230.4 ENST00000494050.1 |
CEP97 |
centrosomal protein 97kDa |
| chr17_-_66287257 | 0.03 |
ENST00000327268.4 |
SLC16A6 |
solute carrier family 16, member 6 |
| chr1_+_176432298 | 0.03 |
ENST00000367661.3 ENST00000367662.3 |
PAPPA2 |
pappalysin 2 |
| chr7_-_229557 | 0.02 |
ENST00000514988.1 |
AC145676.2 |
Uncharacterized protein |
| chr6_+_30594619 | 0.02 |
ENST00000318999.7 ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1 |
alpha tubulin acetyltransferase 1 |
| chr2_-_207024233 | 0.02 |
ENST00000423725.1 ENST00000233190.6 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_+_2398876 | 0.02 |
ENST00000449969.1 |
PLCH2 |
phospholipase C, eta 2 |
| chrX_+_2746850 | 0.02 |
ENST00000381163.3 ENST00000338623.5 ENST00000542787.1 |
GYG2 |
glycogenin 2 |
| chr3_-_30936153 | 0.02 |
ENST00000454381.3 ENST00000282538.5 |
GADL1 |
glutamate decarboxylase-like 1 |
| chr15_+_36887069 | 0.02 |
ENST00000566807.1 ENST00000567389.1 ENST00000562877.1 |
C15orf41 |
chromosome 15 open reading frame 41 |
| chr19_+_4153598 | 0.02 |
ENST00000078445.2 ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3 |
cAMP responsive element binding protein 3-like 3 |
| chr6_+_31895467 | 0.02 |
ENST00000556679.1 ENST00000456570.1 |
CFB CFB |
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chrX_-_132095419 | 0.02 |
ENST00000370836.2 ENST00000521489.1 |
HS6ST2 |
heparan sulfate 6-O-sulfotransferase 2 |
| chr1_-_33815486 | 0.02 |
ENST00000373418.3 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
| chr3_+_184055240 | 0.02 |
ENST00000383847.2 |
FAM131A |
family with sequence similarity 131, member A |
| chr2_-_178257401 | 0.01 |
ENST00000464747.1 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
| chr9_+_1051481 | 0.01 |
ENST00000358146.2 ENST00000259622.6 |
DMRT2 |
doublesex and mab-3 related transcription factor 2 |
| chr18_-_60986962 | 0.01 |
ENST00000333681.4 |
BCL2 |
B-cell CLL/lymphoma 2 |
| chr11_-_8680383 | 0.01 |
ENST00000299550.6 |
TRIM66 |
tripartite motif containing 66 |
| chr14_-_101295407 | 0.01 |
ENST00000596284.1 |
AL117190.2 |
AL117190.2 |
| chr7_+_120969045 | 0.01 |
ENST00000222462.2 |
WNT16 |
wingless-type MMTV integration site family, member 16 |
| chr10_-_75634260 | 0.01 |
ENST00000372765.1 ENST00000351293.3 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
| chr7_-_107443652 | 0.01 |
ENST00000340010.5 ENST00000422236.2 ENST00000453332.1 |
SLC26A3 |
solute carrier family 26 (anion exchanger), member 3 |
| chr11_+_86748998 | 0.01 |
ENST00000525018.1 ENST00000355734.4 |
TMEM135 |
transmembrane protein 135 |
| chr11_+_64073699 | 0.01 |
ENST00000405666.1 ENST00000468670.1 |
ESRRA |
estrogen-related receptor alpha |
| chr14_+_35591735 | 0.01 |
ENST00000604948.1 ENST00000605201.1 ENST00000250377.7 ENST00000321130.10 ENST00000534898.4 |
KIAA0391 |
KIAA0391 |
| chr5_+_73109339 | 0.00 |
ENST00000296799.4 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr3_+_184037466 | 0.00 |
ENST00000441154.1 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr10_-_75634326 | 0.00 |
ENST00000322635.3 ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
| chr3_+_184038073 | 0.00 |
ENST00000428387.1 ENST00000434061.2 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr17_+_41150793 | 0.00 |
ENST00000586277.1 |
RPL27 |
ribosomal protein L27 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060621 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.9 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.3 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.5 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.4 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.1 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.3 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.0 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.0 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 0.0 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.2 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.0 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |