Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
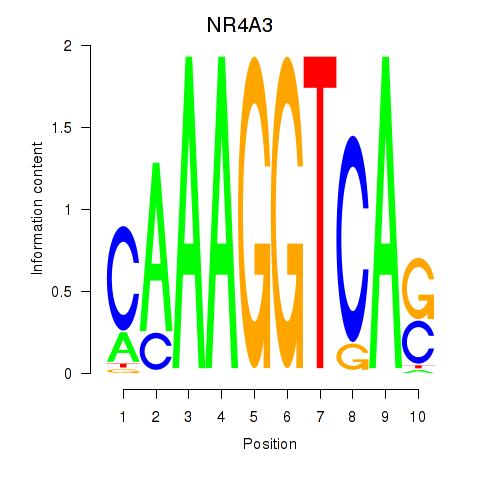
Results for NR4A3
Z-value: 0.44
Transcription factors associated with NR4A3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A3
|
ENSG00000119508.13 | NR4A3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A3 | hg19_v2_chr9_+_102584128_102584144 | 0.61 | 1.1e-01 | Click! |
Activity profile of NR4A3 motif
Sorted Z-values of NR4A3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_31637560 | 0.57 |
ENST00000379416.3 |
XDH |
xanthine dehydrogenase |
| chr10_-_105845674 | 0.51 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr17_-_39677971 | 0.50 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr8_+_86376081 | 0.45 |
ENST00000285379.5 |
CA2 |
carbonic anhydrase II |
| chr1_-_175162048 | 0.42 |
ENST00000444639.1 |
KIAA0040 |
KIAA0040 |
| chr16_+_68679193 | 0.41 |
ENST00000581171.1 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr6_-_11807277 | 0.40 |
ENST00000379415.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr1_+_44401479 | 0.39 |
ENST00000438616.3 |
ARTN |
artemin |
| chr20_+_58179582 | 0.33 |
ENST00000371015.1 ENST00000395639.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr8_+_99956759 | 0.32 |
ENST00000522510.1 ENST00000457907.2 |
OSR2 |
odd-skipped related transciption factor 2 |
| chr15_-_83621435 | 0.29 |
ENST00000450735.2 ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2 |
homer homolog 2 (Drosophila) |
| chr1_+_152957707 | 0.29 |
ENST00000368762.1 |
SPRR1A |
small proline-rich protein 1A |
| chr3_-_151034734 | 0.26 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr22_+_21128167 | 0.26 |
ENST00000215727.5 |
SERPIND1 |
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr1_+_35247859 | 0.25 |
ENST00000373362.3 |
GJB3 |
gap junction protein, beta 3, 31kDa |
| chr5_+_147258266 | 0.25 |
ENST00000296694.4 |
SCGB3A2 |
secretoglobin, family 3A, member 2 |
| chr2_+_17721920 | 0.25 |
ENST00000295156.4 |
VSNL1 |
visinin-like 1 |
| chr10_-_116164450 | 0.24 |
ENST00000369271.3 |
AFAP1L2 |
actin filament associated protein 1-like 2 |
| chr20_+_1875942 | 0.23 |
ENST00000358771.4 |
SIRPA |
signal-regulatory protein alpha |
| chr13_-_52585547 | 0.23 |
ENST00000448424.2 ENST00000400370.3 ENST00000418097.2 ENST00000242839.4 ENST00000400366.3 ENST00000344297.5 |
ATP7B |
ATPase, Cu++ transporting, beta polypeptide |
| chr7_-_20256965 | 0.23 |
ENST00000400331.5 ENST00000332878.4 |
MACC1 |
metastasis associated in colon cancer 1 |
| chr10_-_116164239 | 0.23 |
ENST00000419268.1 ENST00000304129.4 ENST00000545353.1 |
AFAP1L2 |
actin filament associated protein 1-like 2 |
| chr2_+_220492116 | 0.22 |
ENST00000373760.2 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr12_-_54779511 | 0.22 |
ENST00000551109.1 ENST00000546970.1 |
ZNF385A |
zinc finger protein 385A |
| chr1_+_158149737 | 0.22 |
ENST00000368171.3 |
CD1D |
CD1d molecule |
| chr14_-_21492251 | 0.22 |
ENST00000554398.1 |
NDRG2 |
NDRG family member 2 |
| chr20_-_46415297 | 0.22 |
ENST00000467815.1 ENST00000359930.4 |
SULF2 |
sulfatase 2 |
| chr14_-_21492113 | 0.21 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr15_+_90728145 | 0.21 |
ENST00000561085.1 ENST00000379122.3 ENST00000332496.6 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr10_-_116444371 | 0.21 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr20_-_46415341 | 0.20 |
ENST00000484875.1 ENST00000361612.4 |
SULF2 |
sulfatase 2 |
| chr10_-_76995675 | 0.20 |
ENST00000469299.1 |
COMTD1 |
catechol-O-methyltransferase domain containing 1 |
| chr17_-_39928106 | 0.20 |
ENST00000540235.1 |
JUP |
junction plakoglobin |
| chr3_-_197300194 | 0.19 |
ENST00000358186.2 ENST00000431056.1 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
| chr16_+_691792 | 0.19 |
ENST00000307650.4 |
FAM195A |
family with sequence similarity 195, member A |
| chr15_+_45422178 | 0.19 |
ENST00000389037.3 ENST00000558322.1 |
DUOX1 |
dual oxidase 1 |
| chr11_+_66824276 | 0.19 |
ENST00000308831.2 |
RHOD |
ras homolog family member D |
| chr15_+_45422131 | 0.19 |
ENST00000321429.4 |
DUOX1 |
dual oxidase 1 |
| chr10_-_98031265 | 0.18 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr7_-_16840820 | 0.18 |
ENST00000450569.1 |
AGR2 |
anterior gradient 2 |
| chr18_-_67873078 | 0.18 |
ENST00000255674.6 |
RTTN |
rotatin |
| chr8_+_145202939 | 0.18 |
ENST00000423230.2 ENST00000398656.4 |
MROH1 |
maestro heat-like repeat family member 1 |
| chr2_-_238499725 | 0.18 |
ENST00000264601.3 |
RAB17 |
RAB17, member RAS oncogene family |
| chr10_+_81107271 | 0.18 |
ENST00000448165.1 |
PPIF |
peptidylprolyl isomerase F |
| chr12_+_113376157 | 0.18 |
ENST00000228928.7 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr10_-_98031310 | 0.18 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chrX_-_70326455 | 0.17 |
ENST00000374251.5 |
CXorf65 |
chromosome X open reading frame 65 |
| chr17_+_37784749 | 0.17 |
ENST00000394265.1 ENST00000394267.2 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr2_-_238499337 | 0.17 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr17_-_39274606 | 0.17 |
ENST00000391413.2 |
KRTAP4-11 |
keratin associated protein 4-11 |
| chr2_+_220491973 | 0.17 |
ENST00000358055.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr22_+_45072925 | 0.17 |
ENST00000006251.7 |
PRR5 |
proline rich 5 (renal) |
| chr18_+_11981547 | 0.17 |
ENST00000588927.1 |
IMPA2 |
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr22_+_45072958 | 0.17 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr10_-_116286563 | 0.17 |
ENST00000369253.2 |
ABLIM1 |
actin binding LIM protein 1 |
| chr6_-_43197189 | 0.16 |
ENST00000509253.1 ENST00000393987.2 ENST00000230431.6 |
DNPH1 |
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr4_-_40517984 | 0.16 |
ENST00000381795.6 |
RBM47 |
RNA binding motif protein 47 |
| chr1_+_26869597 | 0.16 |
ENST00000530003.1 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr11_+_10472223 | 0.16 |
ENST00000396554.3 ENST00000524866.1 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr1_-_185597619 | 0.15 |
ENST00000608417.1 ENST00000436955.1 |
GS1-204I12.1 |
GS1-204I12.1 |
| chr19_+_34287174 | 0.15 |
ENST00000587559.1 ENST00000588637.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr3_+_113616317 | 0.15 |
ENST00000440446.2 ENST00000488680.1 |
GRAMD1C |
GRAM domain containing 1C |
| chr7_-_139763521 | 0.15 |
ENST00000263549.3 |
PARP12 |
poly (ADP-ribose) polymerase family, member 12 |
| chr2_-_238499131 | 0.15 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr19_-_38743878 | 0.15 |
ENST00000587515.1 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr11_+_66824346 | 0.15 |
ENST00000532559.1 |
RHOD |
ras homolog family member D |
| chr17_-_7991021 | 0.15 |
ENST00000319144.4 |
ALOX12B |
arachidonate 12-lipoxygenase, 12R type |
| chr1_-_183559693 | 0.14 |
ENST00000367535.3 ENST00000413720.1 ENST00000418089.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr2_-_238499303 | 0.14 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr1_+_92495528 | 0.14 |
ENST00000370383.4 |
EPHX4 |
epoxide hydrolase 4 |
| chr1_+_196857144 | 0.14 |
ENST00000367416.2 ENST00000251424.4 ENST00000367418.2 |
CFHR4 |
complement factor H-related 4 |
| chr6_-_33041378 | 0.14 |
ENST00000428995.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr2_+_37458928 | 0.14 |
ENST00000439218.1 ENST00000432075.1 |
NDUFAF7 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr1_+_24645807 | 0.14 |
ENST00000361548.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr11_+_10476851 | 0.14 |
ENST00000396553.2 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr18_+_55816546 | 0.14 |
ENST00000435432.2 ENST00000357895.5 ENST00000586263.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr16_+_57653989 | 0.13 |
ENST00000567835.1 ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr2_-_75796837 | 0.13 |
ENST00000233712.1 |
EVA1A |
eva-1 homolog A (C. elegans) |
| chr14_-_106845789 | 0.13 |
ENST00000390617.2 |
IGHV3-35 |
immunoglobulin heavy variable 3-35 (non-functional) |
| chr10_-_118502070 | 0.13 |
ENST00000369209.3 |
HSPA12A |
heat shock 70kDa protein 12A |
| chr8_+_99956662 | 0.13 |
ENST00000523368.1 ENST00000297565.4 ENST00000435298.2 |
OSR2 |
odd-skipped related transciption factor 2 |
| chr20_+_61287711 | 0.13 |
ENST00000370507.1 |
SLCO4A1 |
solute carrier organic anion transporter family, member 4A1 |
| chr6_-_170599561 | 0.13 |
ENST00000366756.3 |
DLL1 |
delta-like 1 (Drosophila) |
| chr1_+_24646002 | 0.13 |
ENST00000356046.2 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr10_-_76995769 | 0.13 |
ENST00000372538.3 |
COMTD1 |
catechol-O-methyltransferase domain containing 1 |
| chr1_+_24645865 | 0.13 |
ENST00000342072.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr5_+_175085033 | 0.12 |
ENST00000377291.2 |
HRH2 |
histamine receptor H2 |
| chr1_-_116311402 | 0.12 |
ENST00000261448.5 |
CASQ2 |
calsequestrin 2 (cardiac muscle) |
| chr1_-_1356719 | 0.12 |
ENST00000520296.1 |
ANKRD65 |
ankyrin repeat domain 65 |
| chr7_+_121513143 | 0.12 |
ENST00000393386.2 |
PTPRZ1 |
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr4_+_24797085 | 0.12 |
ENST00000382120.3 |
SOD3 |
superoxide dismutase 3, extracellular |
| chr7_+_117864708 | 0.12 |
ENST00000357099.4 ENST00000265224.4 ENST00000486422.1 ENST00000417525.1 |
ANKRD7 |
ankyrin repeat domain 7 |
| chr8_-_131028660 | 0.12 |
ENST00000401979.2 ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr10_-_8095412 | 0.12 |
ENST00000458727.1 ENST00000355358.1 |
RP11-379F12.3 GATA3-AS1 |
RP11-379F12.3 GATA3 antisense RNA 1 |
| chrX_-_43741594 | 0.12 |
ENST00000536181.1 ENST00000378069.4 |
MAOB |
monoamine oxidase B |
| chr12_-_8088871 | 0.12 |
ENST00000075120.7 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chrX_+_56259316 | 0.12 |
ENST00000468660.1 |
KLF8 |
Kruppel-like factor 8 |
| chr17_+_32646055 | 0.11 |
ENST00000394620.1 |
CCL8 |
chemokine (C-C motif) ligand 8 |
| chr3_-_190167571 | 0.11 |
ENST00000354905.2 |
TMEM207 |
transmembrane protein 207 |
| chr11_-_66496430 | 0.11 |
ENST00000533211.1 |
SPTBN2 |
spectrin, beta, non-erythrocytic 2 |
| chr17_-_6616678 | 0.11 |
ENST00000381074.4 ENST00000293800.6 ENST00000572352.1 ENST00000576323.1 ENST00000573648.1 |
SLC13A5 |
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr16_-_20709066 | 0.11 |
ENST00000520010.1 |
ACSM1 |
acyl-CoA synthetase medium-chain family member 1 |
| chr17_+_1646130 | 0.11 |
ENST00000453066.1 ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr18_+_55888767 | 0.11 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr12_-_95510743 | 0.11 |
ENST00000551521.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr4_+_41614909 | 0.11 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr11_+_67033881 | 0.11 |
ENST00000308595.5 ENST00000526285.1 |
ADRBK1 |
adrenergic, beta, receptor kinase 1 |
| chrX_+_65384182 | 0.11 |
ENST00000441993.2 ENST00000419594.1 |
HEPH |
hephaestin |
| chr4_+_6988884 | 0.11 |
ENST00000451522.2 |
TBC1D14 |
TBC1 domain family, member 14 |
| chr1_-_15911510 | 0.11 |
ENST00000375826.3 |
AGMAT |
agmatine ureohydrolase (agmatinase) |
| chr9_+_124103625 | 0.10 |
ENST00000594963.1 |
AL161784.1 |
Uncharacterized protein |
| chr3_-_58652523 | 0.10 |
ENST00000489857.1 ENST00000358781.2 |
FAM3D |
family with sequence similarity 3, member D |
| chr16_+_57653854 | 0.10 |
ENST00000568908.1 ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr1_-_1356628 | 0.10 |
ENST00000442470.1 ENST00000537107.1 |
ANKRD65 |
ankyrin repeat domain 65 |
| chr11_-_123756334 | 0.10 |
ENST00000528595.1 ENST00000375026.2 |
TMEM225 |
transmembrane protein 225 |
| chr17_-_7307358 | 0.10 |
ENST00000576017.1 ENST00000302422.3 ENST00000535512.1 |
TMEM256 TMEM256-PLSCR3 |
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr6_+_18155632 | 0.10 |
ENST00000297792.5 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chr2_+_220325977 | 0.10 |
ENST00000396686.1 ENST00000396689.2 |
SPEG |
SPEG complex locus |
| chr19_-_44143939 | 0.10 |
ENST00000222374.2 |
CADM4 |
cell adhesion molecule 4 |
| chr1_-_152552980 | 0.10 |
ENST00000368787.3 |
LCE3D |
late cornified envelope 3D |
| chr15_+_59730348 | 0.10 |
ENST00000288228.5 ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A |
family with sequence similarity 81, member A |
| chr1_-_111148969 | 0.10 |
ENST00000316361.4 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr3_+_44916098 | 0.10 |
ENST00000296125.4 |
TGM4 |
transglutaminase 4 |
| chr17_+_55173933 | 0.09 |
ENST00000539273.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr7_-_151433342 | 0.09 |
ENST00000433631.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr1_+_24117693 | 0.09 |
ENST00000374503.3 ENST00000374502.3 |
LYPLA2 |
lysophospholipase II |
| chr18_-_6929829 | 0.09 |
ENST00000583840.1 ENST00000581571.1 ENST00000578497.1 ENST00000579012.1 |
LINC00668 |
long intergenic non-protein coding RNA 668 |
| chr14_-_106622419 | 0.09 |
ENST00000390604.2 |
IGHV3-16 |
immunoglobulin heavy variable 3-16 (non-functional) |
| chr1_-_2461684 | 0.09 |
ENST00000378453.3 |
HES5 |
hes family bHLH transcription factor 5 |
| chrX_+_65384052 | 0.09 |
ENST00000336279.5 ENST00000458621.1 |
HEPH |
hephaestin |
| chr3_+_158288942 | 0.09 |
ENST00000491767.1 ENST00000355893.5 |
MLF1 |
myeloid leukemia factor 1 |
| chr2_+_234686976 | 0.09 |
ENST00000389758.3 |
MROH2A |
maestro heat-like repeat family member 2A |
| chrX_+_23801280 | 0.09 |
ENST00000379251.3 ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1 |
spermidine/spermine N1-acetyltransferase 1 |
| chr14_-_21491477 | 0.09 |
ENST00000298684.5 ENST00000557169.1 ENST00000553563.1 |
NDRG2 |
NDRG family member 2 |
| chr11_-_77734260 | 0.09 |
ENST00000353172.5 |
KCTD14 |
potassium channel tetramerization domain containing 14 |
| chr6_-_3457256 | 0.09 |
ENST00000436008.2 |
SLC22A23 |
solute carrier family 22, member 23 |
| chr14_+_77648167 | 0.09 |
ENST00000554346.1 ENST00000298351.4 |
TMEM63C |
transmembrane protein 63C |
| chr4_-_120548779 | 0.09 |
ENST00000264805.5 |
PDE5A |
phosphodiesterase 5A, cGMP-specific |
| chr2_+_27237615 | 0.09 |
ENST00000458529.1 ENST00000402218.1 |
MAPRE3 |
microtubule-associated protein, RP/EB family, member 3 |
| chr11_+_63137251 | 0.09 |
ENST00000310969.4 ENST00000279178.3 |
SLC22A9 |
solute carrier family 22 (organic anion transporter), member 9 |
| chr1_-_46769261 | 0.09 |
ENST00000343304.6 |
LRRC41 |
leucine rich repeat containing 41 |
| chr2_+_88991162 | 0.09 |
ENST00000283646.4 |
RPIA |
ribose 5-phosphate isomerase A |
| chr10_+_101542462 | 0.09 |
ENST00000370449.4 ENST00000370434.1 |
ABCC2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr16_+_85942594 | 0.09 |
ENST00000566369.1 |
IRF8 |
interferon regulatory factor 8 |
| chr4_+_155484155 | 0.09 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr18_-_6929797 | 0.09 |
ENST00000581725.1 ENST00000583316.1 |
LINC00668 |
long intergenic non-protein coding RNA 668 |
| chr15_+_58724184 | 0.09 |
ENST00000433326.2 |
LIPC |
lipase, hepatic |
| chr11_-_74442430 | 0.08 |
ENST00000376332.3 |
CHRDL2 |
chordin-like 2 |
| chr17_+_79761997 | 0.08 |
ENST00000400723.3 ENST00000570996.1 |
GCGR |
glucagon receptor |
| chr1_+_24117627 | 0.08 |
ENST00000400061.1 |
LYPLA2 |
lysophospholipase II |
| chr16_+_31539183 | 0.08 |
ENST00000302312.4 |
AHSP |
alpha hemoglobin stabilizing protein |
| chr11_-_117698765 | 0.08 |
ENST00000532119.1 |
FXYD2 |
FXYD domain containing ion transport regulator 2 |
| chr18_+_56530794 | 0.08 |
ENST00000590285.1 ENST00000586085.1 ENST00000589288.1 |
ZNF532 |
zinc finger protein 532 |
| chr20_-_62130474 | 0.08 |
ENST00000217182.3 |
EEF1A2 |
eukaryotic translation elongation factor 1 alpha 2 |
| chr11_+_35198118 | 0.08 |
ENST00000525211.1 ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr7_-_151433393 | 0.08 |
ENST00000492843.1 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr19_-_11639910 | 0.08 |
ENST00000588998.1 ENST00000586149.1 |
ECSIT |
ECSIT signalling integrator |
| chr16_+_11038403 | 0.08 |
ENST00000409552.3 |
CLEC16A |
C-type lectin domain family 16, member A |
| chr20_-_35807741 | 0.08 |
ENST00000434295.1 ENST00000441008.2 ENST00000400441.3 ENST00000343811.4 |
MROH8 |
maestro heat-like repeat family member 8 |
| chr1_-_111148241 | 0.08 |
ENST00000440270.1 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr7_-_38305279 | 0.08 |
ENST00000443402.2 |
TRGC1 |
T cell receptor gamma constant 1 |
| chr22_+_29469012 | 0.08 |
ENST00000400335.4 ENST00000400338.2 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chr14_+_32546274 | 0.08 |
ENST00000396582.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr3_+_158288960 | 0.08 |
ENST00000484955.1 ENST00000359117.5 ENST00000498592.1 ENST00000477042.1 ENST00000471745.1 ENST00000469452.1 |
MLF1 |
myeloid leukemia factor 1 |
| chr13_-_95364389 | 0.08 |
ENST00000376945.2 |
SOX21 |
SRY (sex determining region Y)-box 21 |
| chr14_+_32546145 | 0.08 |
ENST00000556611.1 ENST00000539826.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr16_+_83932684 | 0.08 |
ENST00000262430.4 |
MLYCD |
malonyl-CoA decarboxylase |
| chr1_-_27701307 | 0.08 |
ENST00000270879.4 ENST00000354982.2 |
FCN3 |
ficolin (collagen/fibrinogen domain containing) 3 |
| chr15_-_99791413 | 0.07 |
ENST00000394129.2 ENST00000558663.1 ENST00000394135.3 ENST00000561365.1 ENST00000560279.1 |
TTC23 |
tetratricopeptide repeat domain 23 |
| chr22_+_46546494 | 0.07 |
ENST00000396000.2 ENST00000262735.5 ENST00000420804.1 |
PPARA |
peroxisome proliferator-activated receptor alpha |
| chr1_+_46769303 | 0.07 |
ENST00000311672.5 |
UQCRH |
ubiquinol-cytochrome c reductase hinge protein |
| chr19_-_48867291 | 0.07 |
ENST00000435956.3 |
TMEM143 |
transmembrane protein 143 |
| chr19_-_42927251 | 0.07 |
ENST00000597001.1 |
LIPE |
lipase, hormone-sensitive |
| chr1_-_36916066 | 0.07 |
ENST00000315643.9 |
OSCP1 |
organic solute carrier partner 1 |
| chrX_+_72223352 | 0.07 |
ENST00000373521.2 ENST00000538388.1 |
PABPC1L2B |
poly(A) binding protein, cytoplasmic 1-like 2B |
| chr12_+_50898881 | 0.07 |
ENST00000301180.5 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr19_-_11639931 | 0.07 |
ENST00000592312.1 ENST00000590480.1 ENST00000585318.1 ENST00000252440.7 ENST00000417981.2 ENST00000270517.7 |
ECSIT |
ECSIT signalling integrator |
| chr22_+_30821732 | 0.07 |
ENST00000355143.4 |
MTFP1 |
mitochondrial fission process 1 |
| chr17_-_40264692 | 0.07 |
ENST00000591220.1 ENST00000251642.3 |
DHX58 |
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr1_+_64059332 | 0.07 |
ENST00000540265.1 |
PGM1 |
phosphoglucomutase 1 |
| chr19_-_48867171 | 0.07 |
ENST00000377431.2 ENST00000436660.2 ENST00000541566.1 |
TMEM143 |
transmembrane protein 143 |
| chr3_-_156840776 | 0.07 |
ENST00000471357.1 |
LINC00880 |
long intergenic non-protein coding RNA 880 |
| chr5_+_1801503 | 0.07 |
ENST00000274137.5 ENST00000469176.1 |
NDUFS6 |
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr1_-_151138323 | 0.07 |
ENST00000368908.5 |
LYSMD1 |
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr7_-_37956409 | 0.07 |
ENST00000436072.2 |
SFRP4 |
secreted frizzled-related protein 4 |
| chr14_-_106209368 | 0.07 |
ENST00000390548.2 ENST00000390549.2 ENST00000390542.2 |
IGHG1 |
immunoglobulin heavy constant gamma 1 (G1m marker) |
| chr3_+_186435137 | 0.07 |
ENST00000447445.1 |
KNG1 |
kininogen 1 |
| chr1_-_36947120 | 0.07 |
ENST00000361632.4 |
CSF3R |
colony stimulating factor 3 receptor (granulocyte) |
| chr8_-_131028869 | 0.07 |
ENST00000518283.1 ENST00000519110.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr17_+_40440481 | 0.07 |
ENST00000590726.2 ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A |
signal transducer and activator of transcription 5A |
| chr8_-_70745575 | 0.07 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr19_+_39616410 | 0.07 |
ENST00000602004.1 ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4 |
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr19_-_633576 | 0.07 |
ENST00000588649.2 |
POLRMT |
polymerase (RNA) mitochondrial (DNA directed) |
| chr12_-_109219937 | 0.07 |
ENST00000546697.1 |
SSH1 |
slingshot protein phosphatase 1 |
| chr22_-_29711645 | 0.07 |
ENST00000401450.3 |
RASL10A |
RAS-like, family 10, member A |
| chr19_+_14138960 | 0.07 |
ENST00000431365.2 ENST00000585987.1 |
RLN3 |
relaxin 3 |
| chr12_+_70219052 | 0.07 |
ENST00000552032.2 ENST00000547771.2 |
MYRFL |
myelin regulatory factor-like |
| chr10_+_81272287 | 0.07 |
ENST00000520547.2 |
EIF5AL1 |
eukaryotic translation initiation factor 5A-like 1 |
| chrX_+_47078069 | 0.07 |
ENST00000357227.4 ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16 |
cyclin-dependent kinase 16 |
| chr22_+_20104947 | 0.07 |
ENST00000402752.1 |
RANBP1 |
RAN binding protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.6 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.4 | GO:0032773 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.1 | 0.6 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.5 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 0.4 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.2 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.1 | 0.4 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.2 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.2 | GO:2000981 | negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.2 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.5 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.3 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.2 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.3 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.0 | GO:0042323 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.0 | 0.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) putrescine biosynthetic process(GO:0009446) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.4 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:2000366 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.0 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.1 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0010044 | response to aluminum ion(GO:0010044) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.1 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.1 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.0 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0071415 | cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.0 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.0 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.5 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.6 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.2 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.1 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.2 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.3 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.0 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |