Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for NR6A1
Z-value: 0.32
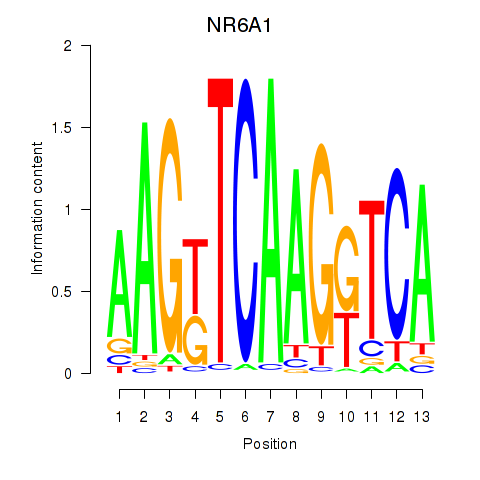
Transcription factors associated with NR6A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR6A1
|
ENSG00000148200.12 | NR6A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR6A1 | hg19_v2_chr9_-_127533519_127533576 | -0.63 | 9.7e-02 | Click! |
Activity profile of NR6A1 motif
Sorted Z-values of NR6A1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR6A1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_84743436 | 0.46 |
ENST00000257776.4 |
MRAP2 |
melanocortin 2 receptor accessory protein 2 |
| chr12_-_91574142 | 0.42 |
ENST00000547937.1 |
DCN |
decorin |
| chr1_-_153517473 | 0.35 |
ENST00000368715.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr6_-_31697977 | 0.31 |
ENST00000375787.2 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr16_+_77756399 | 0.29 |
ENST00000564085.1 ENST00000268533.5 ENST00000568787.1 ENST00000437314.3 ENST00000563839.1 |
NUDT7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chrX_+_22050546 | 0.28 |
ENST00000379374.4 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr19_-_59023348 | 0.26 |
ENST00000601355.1 ENST00000263093.2 |
SLC27A5 |
solute carrier family 27 (fatty acid transporter), member 5 |
| chr4_-_159094194 | 0.25 |
ENST00000592057.1 ENST00000585682.1 ENST00000393807.5 |
FAM198B |
family with sequence similarity 198, member B |
| chr10_+_89419370 | 0.25 |
ENST00000361175.4 ENST00000456849.1 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr5_-_14871866 | 0.25 |
ENST00000284268.6 |
ANKH |
ANKH inorganic pyrophosphate transport regulator |
| chr1_+_109102652 | 0.23 |
ENST00000370035.3 ENST00000405454.1 |
FAM102B |
family with sequence similarity 102, member B |
| chr20_-_30310336 | 0.23 |
ENST00000434194.1 ENST00000376062.2 |
BCL2L1 |
BCL2-like 1 |
| chr6_-_43197189 | 0.23 |
ENST00000509253.1 ENST00000393987.2 ENST00000230431.6 |
DNPH1 |
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr8_-_27472198 | 0.23 |
ENST00000519472.1 ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU |
clusterin |
| chrX_-_118827333 | 0.23 |
ENST00000360156.7 ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6 |
septin 6 |
| chr3_-_58613323 | 0.22 |
ENST00000474531.1 ENST00000465970.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr7_-_130353553 | 0.22 |
ENST00000330992.7 ENST00000445977.2 |
COPG2 |
coatomer protein complex, subunit gamma 2 |
| chr20_-_48099182 | 0.21 |
ENST00000371741.4 |
KCNB1 |
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr19_-_58864848 | 0.21 |
ENST00000263100.3 |
A1BG |
alpha-1-B glycoprotein |
| chr15_+_41057818 | 0.20 |
ENST00000558467.1 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr11_+_65190245 | 0.20 |
ENST00000499732.1 ENST00000501122.2 ENST00000601801.1 |
NEAT1 |
nuclear paraspeckle assembly transcript 1 (non-protein coding) |
| chr14_-_90085458 | 0.20 |
ENST00000345097.4 ENST00000555855.1 ENST00000555353.1 |
FOXN3 |
forkhead box N3 |
| chr19_-_10446449 | 0.20 |
ENST00000592439.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr19_+_11455900 | 0.19 |
ENST00000588790.1 |
CCDC159 |
coiled-coil domain containing 159 |
| chr13_-_114898016 | 0.19 |
ENST00000542651.1 ENST00000334062.7 |
RASA3 |
RAS p21 protein activator 3 |
| chr16_-_18801643 | 0.18 |
ENST00000322989.4 ENST00000563390.1 |
RPS15A |
ribosomal protein S15a |
| chr1_-_161102367 | 0.18 |
ENST00000464113.1 |
DEDD |
death effector domain containing |
| chr4_+_126237554 | 0.18 |
ENST00000394329.3 |
FAT4 |
FAT atypical cadherin 4 |
| chrX_+_103031758 | 0.18 |
ENST00000303958.2 ENST00000361621.2 |
PLP1 |
proteolipid protein 1 |
| chr1_+_225997791 | 0.18 |
ENST00000445856.1 ENST00000272167.5 |
EPHX1 |
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr1_+_151739131 | 0.17 |
ENST00000400999.1 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr2_-_175711133 | 0.16 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr3_-_114790179 | 0.16 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chrX_+_103031421 | 0.16 |
ENST00000433491.1 ENST00000418604.1 ENST00000443502.1 |
PLP1 |
proteolipid protein 1 |
| chr11_+_30344595 | 0.16 |
ENST00000282032.3 |
ARL14EP |
ADP-ribosylation factor-like 14 effector protein |
| chr14_-_106539557 | 0.16 |
ENST00000390599.2 |
IGHV1-8 |
immunoglobulin heavy variable 1-8 |
| chr6_-_160166218 | 0.16 |
ENST00000537657.1 |
SOD2 |
superoxide dismutase 2, mitochondrial |
| chr17_+_7452189 | 0.16 |
ENST00000293825.6 |
TNFSF12 |
tumor necrosis factor (ligand) superfamily, member 12 |
| chr7_+_43803790 | 0.16 |
ENST00000424330.1 |
BLVRA |
biliverdin reductase A |
| chr20_+_33462888 | 0.16 |
ENST00000336325.4 |
ACSS2 |
acyl-CoA synthetase short-chain family member 2 |
| chrX_+_102840408 | 0.15 |
ENST00000468024.1 ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4 |
transcription elongation factor A (SII)-like 4 |
| chr19_+_10196981 | 0.15 |
ENST00000591813.1 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr9_-_21305312 | 0.15 |
ENST00000259555.4 |
IFNA5 |
interferon, alpha 5 |
| chr19_+_42082506 | 0.15 |
ENST00000187608.9 ENST00000401445.2 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
| chr7_-_108168580 | 0.15 |
ENST00000453085.1 |
PNPLA8 |
patatin-like phospholipase domain containing 8 |
| chr9_+_33795533 | 0.14 |
ENST00000379405.3 |
PRSS3 |
protease, serine, 3 |
| chr22_+_30163340 | 0.14 |
ENST00000330029.6 ENST00000401406.3 |
UQCR10 |
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr19_+_17516909 | 0.14 |
ENST00000601007.1 ENST00000594913.1 ENST00000599975.1 ENST00000600514.1 |
CTD-2521M24.9 MVB12A |
CTD-2521M24.9 multivesicular body subunit 12A |
| chr6_+_25652432 | 0.14 |
ENST00000377961.2 |
SCGN |
secretagogin, EF-hand calcium binding protein |
| chr5_-_140027175 | 0.14 |
ENST00000512088.1 |
NDUFA2 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chrX_-_118827113 | 0.14 |
ENST00000394617.2 |
SEPT6 |
septin 6 |
| chr9_+_123970052 | 0.14 |
ENST00000373823.3 |
GSN |
gelsolin |
| chr3_-_156840776 | 0.14 |
ENST00000471357.1 |
LINC00880 |
long intergenic non-protein coding RNA 880 |
| chr8_-_97247759 | 0.14 |
ENST00000518406.1 ENST00000523920.1 ENST00000287022.5 |
UQCRB |
ubiquinol-cytochrome c reductase binding protein |
| chr14_-_106471723 | 0.14 |
ENST00000390595.2 |
IGHV1-3 |
immunoglobulin heavy variable 1-3 |
| chr6_-_43027105 | 0.14 |
ENST00000230413.5 ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2 |
mitochondrial ribosomal protein L2 |
| chr19_+_10197463 | 0.13 |
ENST00000590378.1 ENST00000397881.3 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr7_-_56160625 | 0.13 |
ENST00000446428.1 ENST00000432123.1 ENST00000452681.2 ENST00000537360.1 |
PHKG1 |
phosphorylase kinase, gamma 1 (muscle) |
| chr21_+_33671160 | 0.13 |
ENST00000303645.5 |
MRAP |
melanocortin 2 receptor accessory protein |
| chr8_-_72268889 | 0.13 |
ENST00000388742.4 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr12_+_10366016 | 0.13 |
ENST00000546017.1 ENST00000535576.1 ENST00000539170.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr17_+_66509019 | 0.13 |
ENST00000585981.1 ENST00000589480.1 ENST00000585815.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr5_+_134240800 | 0.13 |
ENST00000512783.1 |
PCBD2 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
| chr7_-_6048650 | 0.13 |
ENST00000382321.4 ENST00000406569.3 |
PMS2 |
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr16_-_420514 | 0.13 |
ENST00000199706.8 |
MRPL28 |
mitochondrial ribosomal protein L28 |
| chr7_-_75401513 | 0.13 |
ENST00000005180.4 |
CCL26 |
chemokine (C-C motif) ligand 26 |
| chr7_-_141957847 | 0.13 |
ENST00000552471.1 ENST00000547058.2 |
PRSS58 |
protease, serine, 58 |
| chr1_+_19923454 | 0.12 |
ENST00000602662.1 ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1 MINOS1 |
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr11_-_64764435 | 0.12 |
ENST00000534177.1 ENST00000301887.4 |
BATF2 |
basic leucine zipper transcription factor, ATF-like 2 |
| chr3_+_51976338 | 0.12 |
ENST00000417220.2 ENST00000431474.1 ENST00000398755.3 |
PARP3 |
poly (ADP-ribose) polymerase family, member 3 |
| chr5_-_140027357 | 0.12 |
ENST00000252102.4 |
NDUFA2 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr1_-_102312517 | 0.12 |
ENST00000338858.5 |
OLFM3 |
olfactomedin 3 |
| chrX_-_46187069 | 0.12 |
ENST00000446884.1 |
RP1-30G7.2 |
RP1-30G7.2 |
| chr5_-_93447333 | 0.11 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr5_-_107717782 | 0.11 |
ENST00000542267.1 |
FBXL17 |
F-box and leucine-rich repeat protein 17 |
| chr19_+_41117770 | 0.11 |
ENST00000601032.1 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr3_-_48754599 | 0.11 |
ENST00000413654.1 ENST00000454335.1 ENST00000440424.1 ENST00000449610.1 ENST00000443964.1 ENST00000417896.1 ENST00000413298.1 ENST00000449563.1 ENST00000443853.1 ENST00000437427.1 ENST00000446860.1 ENST00000412850.1 ENST00000424035.1 ENST00000340879.4 ENST00000431721.2 ENST00000434860.1 ENST00000328631.5 ENST00000432678.2 |
IP6K2 |
inositol hexakisphosphate kinase 2 |
| chr1_-_161102421 | 0.11 |
ENST00000490843.2 ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD |
death effector domain containing |
| chr2_+_203104292 | 0.11 |
ENST00000594829.1 |
AC079354.2 |
Uncharacterized protein |
| chr8_-_72268968 | 0.11 |
ENST00000388740.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr1_-_48866517 | 0.11 |
ENST00000371841.1 |
SPATA6 |
spermatogenesis associated 6 |
| chr3_+_39851094 | 0.11 |
ENST00000302541.6 |
MYRIP |
myosin VIIA and Rab interacting protein |
| chr11_+_65407331 | 0.11 |
ENST00000527525.1 |
SIPA1 |
signal-induced proliferation-associated 1 |
| chr4_+_667307 | 0.11 |
ENST00000506838.1 |
MYL5 |
myosin, light chain 5, regulatory |
| chr15_+_75182346 | 0.11 |
ENST00000569931.1 ENST00000352410.4 ENST00000566377.1 ENST00000569233.1 ENST00000567132.1 ENST00000564633.1 ENST00000568907.1 ENST00000563422.1 ENST00000564003.1 ENST00000562800.1 ENST00000563786.1 ENST00000535694.1 ENST00000323744.6 ENST00000568828.1 ENST00000562606.1 ENST00000565576.1 ENST00000567570.1 |
MPI |
mannose phosphate isomerase |
| chr2_+_26467825 | 0.11 |
ENST00000545822.1 ENST00000425035.1 |
HADHB |
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit |
| chr16_-_12897642 | 0.11 |
ENST00000433677.2 ENST00000261660.4 ENST00000381774.4 |
CPPED1 |
calcineurin-like phosphoesterase domain containing 1 |
| chr7_-_14026063 | 0.10 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr5_+_140749803 | 0.10 |
ENST00000576222.1 |
PCDHGB3 |
protocadherin gamma subfamily B, 3 |
| chr1_-_154928562 | 0.10 |
ENST00000368463.3 ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1 |
pre-B-cell leukemia homeobox interacting protein 1 |
| chr19_-_53770972 | 0.10 |
ENST00000311170.4 |
VN1R4 |
vomeronasal 1 receptor 4 |
| chr19_-_11669960 | 0.10 |
ENST00000589171.1 ENST00000590700.1 ENST00000586683.1 ENST00000593077.1 ENST00000252445.3 |
ELOF1 |
elongation factor 1 homolog (S. cerevisiae) |
| chr15_-_20170354 | 0.10 |
ENST00000338912.5 |
IGHV1OR15-9 |
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr17_-_26903900 | 0.10 |
ENST00000395319.3 ENST00000581807.1 ENST00000584086.1 ENST00000395321.2 |
ALDOC |
aldolase C, fructose-bisphosphate |
| chr10_+_124030819 | 0.10 |
ENST00000260723.4 ENST00000368994.2 |
BTBD16 |
BTB (POZ) domain containing 16 |
| chr7_-_6048702 | 0.10 |
ENST00000265849.7 |
PMS2 |
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr2_+_26467762 | 0.10 |
ENST00000317799.5 ENST00000405867.3 ENST00000537713.1 |
HADHB |
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), beta subunit |
| chr12_-_2027639 | 0.10 |
ENST00000586184.1 ENST00000587995.1 ENST00000585732.1 |
CACNA2D4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr11_-_78052923 | 0.10 |
ENST00000340149.2 |
GAB2 |
GRB2-associated binding protein 2 |
| chr16_+_777118 | 0.10 |
ENST00000562141.1 |
HAGHL |
hydroxyacylglutathione hydrolase-like |
| chr9_-_114362093 | 0.10 |
ENST00000538962.1 ENST00000407693.2 ENST00000238248.3 |
PTGR1 |
prostaglandin reductase 1 |
| chr8_-_27468842 | 0.10 |
ENST00000523500.1 |
CLU |
clusterin |
| chr12_+_26164645 | 0.10 |
ENST00000542004.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr1_-_20126365 | 0.10 |
ENST00000294543.6 ENST00000375122.2 |
TMCO4 |
transmembrane and coiled-coil domains 4 |
| chr3_+_151986709 | 0.10 |
ENST00000495875.2 ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr14_+_74058410 | 0.10 |
ENST00000326303.4 |
ACOT4 |
acyl-CoA thioesterase 4 |
| chr14_-_47120956 | 0.10 |
ENST00000298283.3 |
RPL10L |
ribosomal protein L10-like |
| chr12_+_130554803 | 0.10 |
ENST00000535487.1 |
RP11-474D1.2 |
RP11-474D1.2 |
| chr12_+_123942188 | 0.10 |
ENST00000526639.2 |
SNRNP35 |
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr6_+_31620191 | 0.10 |
ENST00000375918.2 ENST00000375920.4 |
APOM |
apolipoprotein M |
| chr18_+_11751466 | 0.10 |
ENST00000535121.1 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr19_-_43530643 | 0.09 |
ENST00000320078.7 |
PSG11 |
pregnancy specific beta-1-glycoprotein 11 |
| chr6_+_43028182 | 0.09 |
ENST00000394058.1 |
KLC4 |
kinesin light chain 4 |
| chr16_-_420338 | 0.09 |
ENST00000450882.1 ENST00000441883.1 ENST00000447696.1 ENST00000389675.2 |
MRPL28 |
mitochondrial ribosomal protein L28 |
| chr10_-_70092671 | 0.09 |
ENST00000358769.2 ENST00000432941.1 ENST00000495025.2 |
PBLD |
phenazine biosynthesis-like protein domain containing |
| chr12_+_9980069 | 0.09 |
ENST00000354855.3 ENST00000324214.4 ENST00000279544.3 |
KLRF1 |
killer cell lectin-like receptor subfamily F, member 1 |
| chr22_+_42372764 | 0.09 |
ENST00000396426.3 ENST00000406029.1 |
SEPT3 |
septin 3 |
| chr17_-_7531121 | 0.09 |
ENST00000573566.1 ENST00000269298.5 |
SAT2 |
spermidine/spermine N1-acetyltransferase family member 2 |
| chr3_-_72496035 | 0.09 |
ENST00000477973.2 |
RYBP |
RING1 and YY1 binding protein |
| chr19_-_43530600 | 0.09 |
ENST00000598133.1 ENST00000403486.1 ENST00000306322.7 ENST00000401740.1 |
PSG11 |
pregnancy specific beta-1-glycoprotein 11 |
| chr1_-_151798546 | 0.09 |
ENST00000356728.6 |
RORC |
RAR-related orphan receptor C |
| chr2_-_26467465 | 0.09 |
ENST00000457468.2 |
HADHA |
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr1_-_44818599 | 0.09 |
ENST00000537474.1 |
ERI3 |
ERI1 exoribonuclease family member 3 |
| chr11_-_68519026 | 0.09 |
ENST00000255087.5 |
MTL5 |
metallothionein-like 5, testis-specific (tesmin) |
| chr9_-_21187598 | 0.09 |
ENST00000421715.1 |
IFNA4 |
interferon, alpha 4 |
| chrX_+_54466829 | 0.09 |
ENST00000375151.4 |
TSR2 |
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr19_+_49990811 | 0.09 |
ENST00000391857.4 ENST00000467825.2 |
RPL13A |
ribosomal protein L13a |
| chr1_-_203198790 | 0.09 |
ENST00000367229.1 ENST00000255427.3 ENST00000535569.1 |
CHIT1 |
chitinase 1 (chitotriosidase) |
| chr8_-_116680833 | 0.09 |
ENST00000220888.5 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr22_-_30867973 | 0.09 |
ENST00000402286.1 ENST00000401751.1 ENST00000539629.1 ENST00000403066.1 ENST00000215812.4 |
SEC14L3 |
SEC14-like 3 (S. cerevisiae) |
| chr18_+_616711 | 0.08 |
ENST00000579494.1 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr1_+_159557607 | 0.08 |
ENST00000255040.2 |
APCS |
amyloid P component, serum |
| chr6_+_146864829 | 0.08 |
ENST00000367495.3 |
RAB32 |
RAB32, member RAS oncogene family |
| chr17_+_48712206 | 0.08 |
ENST00000427699.1 ENST00000285238.8 |
ABCC3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chrX_+_48620147 | 0.08 |
ENST00000303227.6 |
GLOD5 |
glyoxalase domain containing 5 |
| chr17_+_7905912 | 0.08 |
ENST00000254854.4 |
GUCY2D |
guanylate cyclase 2D, membrane (retina-specific) |
| chr17_+_45810594 | 0.08 |
ENST00000177694.1 |
TBX21 |
T-box 21 |
| chr6_-_25874440 | 0.08 |
ENST00000361703.6 ENST00000397060.4 |
SLC17A3 |
solute carrier family 17 (organic anion transporter), member 3 |
| chr11_+_71935797 | 0.08 |
ENST00000298229.2 ENST00000541756.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr19_+_7985198 | 0.08 |
ENST00000221573.6 ENST00000595637.1 |
SNAPC2 |
small nuclear RNA activating complex, polypeptide 2, 45kDa |
| chr9_-_125590818 | 0.08 |
ENST00000259467.4 |
PDCL |
phosducin-like |
| chr21_+_10862622 | 0.08 |
ENST00000302092.5 ENST00000559480.1 |
IGHV1OR21-1 |
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr13_+_100741269 | 0.08 |
ENST00000376286.4 ENST00000376279.3 ENST00000376285.1 |
PCCA |
propionyl CoA carboxylase, alpha polypeptide |
| chr13_+_23755054 | 0.08 |
ENST00000218867.3 |
SGCG |
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr10_-_25010795 | 0.08 |
ENST00000416305.1 ENST00000376410.2 |
ARHGAP21 |
Rho GTPase activating protein 21 |
| chr2_-_3521518 | 0.08 |
ENST00000382093.5 |
ADI1 |
acireductone dioxygenase 1 |
| chrX_-_8139308 | 0.08 |
ENST00000317103.4 |
VCX2 |
variable charge, X-linked 2 |
| chr1_+_9299895 | 0.08 |
ENST00000602477.1 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr2_-_65593784 | 0.08 |
ENST00000443619.2 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
| chr19_+_49999631 | 0.08 |
ENST00000270625.2 ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11 |
ribosomal protein S11 |
| chr8_-_116680208 | 0.08 |
ENST00000517323.2 ENST00000520276.1 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr8_+_67104323 | 0.08 |
ENST00000518412.1 ENST00000518035.1 ENST00000517725.1 |
LINC00967 |
long intergenic non-protein coding RNA 967 |
| chr19_-_43709703 | 0.08 |
ENST00000599391.1 ENST00000244295.9 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chr9_-_21228221 | 0.08 |
ENST00000413767.2 |
IFNA17 |
interferon, alpha 17 |
| chr2_-_175547571 | 0.08 |
ENST00000409415.3 ENST00000359761.3 ENST00000272746.5 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr11_-_111957451 | 0.08 |
ENST00000504148.2 ENST00000541231.1 |
TIMM8B |
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr2_+_101437487 | 0.07 |
ENST00000427413.1 ENST00000542504.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr1_+_197886461 | 0.07 |
ENST00000367388.3 ENST00000337020.2 ENST00000367387.4 |
LHX9 |
LIM homeobox 9 |
| chr4_-_186732048 | 0.07 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr1_+_41157421 | 0.07 |
ENST00000372654.1 |
NFYC |
nuclear transcription factor Y, gamma |
| chr17_-_15587602 | 0.07 |
ENST00000416464.2 ENST00000578237.1 ENST00000581200.1 |
TRIM16 |
tripartite motif containing 16 |
| chr19_-_43709817 | 0.07 |
ENST00000433626.2 ENST00000405312.3 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chr12_-_7261772 | 0.07 |
ENST00000545280.1 ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL |
complement component 1, r subcomponent-like |
| chr14_+_76071805 | 0.07 |
ENST00000539311.1 |
FLVCR2 |
feline leukemia virus subgroup C cellular receptor family, member 2 |
| chrX_+_72223352 | 0.07 |
ENST00000373521.2 ENST00000538388.1 |
PABPC1L2B |
poly(A) binding protein, cytoplasmic 1-like 2B |
| chr19_-_43383789 | 0.07 |
ENST00000595356.1 |
PSG1 |
pregnancy specific beta-1-glycoprotein 1 |
| chr4_+_159593271 | 0.07 |
ENST00000512251.1 ENST00000511912.1 |
ETFDH |
electron-transferring-flavoprotein dehydrogenase |
| chr3_-_126327398 | 0.07 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr18_+_616672 | 0.07 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr17_+_48912744 | 0.07 |
ENST00000311378.4 |
WFIKKN2 |
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr7_-_43769066 | 0.07 |
ENST00000223336.6 ENST00000310564.6 ENST00000431651.1 ENST00000415798.1 |
COA1 |
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
| chr9_-_14314066 | 0.07 |
ENST00000397575.3 |
NFIB |
nuclear factor I/B |
| chr2_+_201994208 | 0.07 |
ENST00000440180.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr7_-_87936195 | 0.07 |
ENST00000414498.1 ENST00000301959.5 ENST00000380079.4 |
STEAP4 |
STEAP family member 4 |
| chr12_+_53399942 | 0.07 |
ENST00000262056.9 |
EIF4B |
eukaryotic translation initiation factor 4B |
| chr2_-_26467557 | 0.07 |
ENST00000380649.3 |
HADHA |
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chr16_+_20817761 | 0.07 |
ENST00000568046.1 ENST00000261377.6 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr19_+_42381337 | 0.07 |
ENST00000597454.1 ENST00000444740.2 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
| chr7_-_43769051 | 0.07 |
ENST00000395880.3 |
COA1 |
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
| chr4_-_57253587 | 0.07 |
ENST00000513376.1 ENST00000602986.1 ENST00000434343.2 ENST00000451613.1 ENST00000205214.6 ENST00000502617.1 |
AASDH |
aminoadipate-semialdehyde dehydrogenase |
| chr3_+_46618727 | 0.06 |
ENST00000296145.5 |
TDGF1 |
teratocarcinoma-derived growth factor 1 |
| chr12_+_108956355 | 0.06 |
ENST00000535729.1 ENST00000431221.2 ENST00000547005.1 ENST00000311893.9 ENST00000392807.4 ENST00000338291.4 ENST00000539593.1 |
ISCU |
iron-sulfur cluster assembly enzyme |
| chr10_+_114710516 | 0.06 |
ENST00000542695.1 ENST00000346198.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr5_-_41213607 | 0.06 |
ENST00000337836.5 ENST00000433294.1 |
C6 |
complement component 6 |
| chr16_-_20364030 | 0.06 |
ENST00000396134.2 ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD |
uromodulin |
| chr1_+_78511586 | 0.06 |
ENST00000370759.3 |
GIPC2 |
GIPC PDZ domain containing family, member 2 |
| chr9_+_114393634 | 0.06 |
ENST00000556107.1 ENST00000374294.3 |
DNAJC25 DNAJC25-GNG10 |
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr14_+_19553365 | 0.06 |
ENST00000409832.3 |
POTEG |
POTE ankyrin domain family, member G |
| chr3_-_139396560 | 0.06 |
ENST00000514703.1 ENST00000511444.1 |
NMNAT3 |
nicotinamide nucleotide adenylyltransferase 3 |
| chr16_-_20364122 | 0.06 |
ENST00000396138.4 ENST00000577168.1 |
UMOD |
uromodulin |
| chr13_+_23755099 | 0.06 |
ENST00000537476.1 |
SGCG |
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr11_+_63742050 | 0.06 |
ENST00000314133.3 ENST00000535431.1 |
COX8A AP000721.4 |
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr20_-_35402123 | 0.06 |
ENST00000373740.3 ENST00000426836.1 ENST00000373745.3 ENST00000448110.2 ENST00000438549.1 ENST00000447406.1 ENST00000373750.4 ENST00000373734.4 |
DSN1 |
DSN1, MIS12 kinetochore complex component |
| chr13_+_98086445 | 0.06 |
ENST00000245304.4 |
RAP2A |
RAP2A, member of RAS oncogene family |
| chr3_+_180630090 | 0.06 |
ENST00000357559.4 ENST00000305586.7 |
FXR1 |
fragile X mental retardation, autosomal homolog 1 |
| chr15_+_76629064 | 0.06 |
ENST00000290759.4 |
ISL2 |
ISL LIM homeobox 2 |
| chr19_-_43690642 | 0.06 |
ENST00000407356.1 ENST00000407568.1 ENST00000404580.1 ENST00000599812.1 |
PSG5 |
pregnancy specific beta-1-glycoprotein 5 |
| chr10_+_106014468 | 0.06 |
ENST00000369710.4 ENST00000369713.5 ENST00000445155.1 |
GSTO1 |
glutathione S-transferase omega 1 |
| chr6_-_42690312 | 0.06 |
ENST00000230381.5 |
PRPH2 |
peripherin 2 (retinal degeneration, slow) |
| chrX_+_47229982 | 0.06 |
ENST00000377073.3 |
ZNF157 |
zinc finger protein 157 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.2 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 0.3 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.3 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 0.3 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.4 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.2 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.3 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.2 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.1 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 0.1 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0042462 | eye photoreceptor cell differentiation(GO:0001754) eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.3 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.0 | GO:0019254 | amino-acid betaine catabolic process(GO:0006579) carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.2 | GO:0009125 | nucleoside monophosphate catabolic process(GO:0009125) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.0 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.0 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.2 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032173 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.0 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.2 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.2 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.1 | GO:0032440 | 2-alkenal reductase [NAD(P)] activity(GO:0032440) |
| 0.0 | 0.2 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0050610 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.0 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0046790 | complement component C1q binding(GO:0001849) virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |