Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
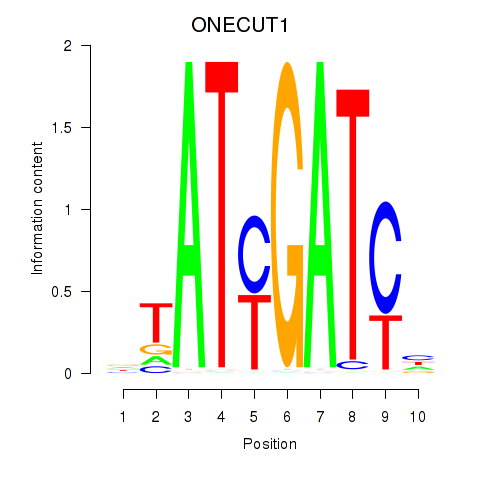
Results for ONECUT1
Z-value: 0.63
Transcription factors associated with ONECUT1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT1
|
ENSG00000169856.7 | ONECUT1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT1 | hg19_v2_chr15_-_53082178_53082209 | -0.16 | 7.0e-01 | Click! |
Activity profile of ONECUT1 motif
Sorted Z-values of ONECUT1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_119799927 | 0.47 |
ENST00000407149.2 ENST00000379551.2 |
PRR16 |
proline rich 16 |
| chr3_+_32280159 | 0.46 |
ENST00000458535.2 ENST00000307526.3 |
CMTM8 |
CKLF-like MARVEL transmembrane domain containing 8 |
| chr5_-_20575959 | 0.42 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr8_-_23712312 | 0.41 |
ENST00000290271.2 |
STC1 |
stanniocalcin 1 |
| chr14_+_101299520 | 0.38 |
ENST00000455531.1 |
MEG3 |
maternally expressed 3 (non-protein coding) |
| chr4_-_157892167 | 0.37 |
ENST00000541126.1 |
PDGFC |
platelet derived growth factor C |
| chr4_-_157892055 | 0.35 |
ENST00000422544.2 |
PDGFC |
platelet derived growth factor C |
| chr12_-_76817036 | 0.32 |
ENST00000546946.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr11_+_19799327 | 0.30 |
ENST00000540292.1 |
NAV2 |
neuron navigator 2 |
| chr8_-_108510224 | 0.28 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr3_+_152017924 | 0.28 |
ENST00000465907.2 ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr3_+_148415571 | 0.28 |
ENST00000497524.1 ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr6_+_73076432 | 0.27 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr4_-_157892498 | 0.27 |
ENST00000502773.1 |
PDGFC |
platelet derived growth factor C |
| chr3_+_69812877 | 0.24 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr1_+_113933371 | 0.23 |
ENST00000369617.4 |
MAGI3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr3_+_191046810 | 0.23 |
ENST00000392455.3 ENST00000392456.3 |
CCDC50 |
coiled-coil domain containing 50 |
| chrX_-_151143140 | 0.22 |
ENST00000393914.3 ENST00000370328.3 ENST00000370325.1 |
GABRE |
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr10_-_4285923 | 0.21 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chr7_-_132261223 | 0.21 |
ENST00000423507.2 |
PLXNA4 |
plexin A4 |
| chr11_-_134123142 | 0.20 |
ENST00000392595.2 ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1 |
thymocyte nuclear protein 1 |
| chr8_-_27468842 | 0.20 |
ENST00000523500.1 |
CLU |
clusterin |
| chr14_+_23012122 | 0.20 |
ENST00000390534.1 |
TRAJ3 |
T cell receptor alpha joining 3 |
| chr11_+_60467047 | 0.20 |
ENST00000300226.2 |
MS4A8 |
membrane-spanning 4-domains, subfamily A, member 8 |
| chr6_-_31651817 | 0.20 |
ENST00000375863.3 ENST00000375860.2 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr1_+_10292308 | 0.19 |
ENST00000377081.1 |
KIF1B |
kinesin family member 1B |
| chr21_+_27011899 | 0.18 |
ENST00000425221.2 |
JAM2 |
junctional adhesion molecule 2 |
| chr5_+_98109322 | 0.18 |
ENST00000513185.1 |
RGMB |
repulsive guidance molecule family member b |
| chr4_-_175443484 | 0.17 |
ENST00000514584.1 ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr12_+_21679220 | 0.17 |
ENST00000256969.2 |
C12orf39 |
chromosome 12 open reading frame 39 |
| chrX_+_10124977 | 0.17 |
ENST00000380833.4 |
CLCN4 |
chloride channel, voltage-sensitive 4 |
| chr15_-_89089860 | 0.16 |
ENST00000558413.1 ENST00000564406.1 ENST00000268148.8 |
DET1 |
de-etiolated homolog 1 (Arabidopsis) |
| chr1_+_145301735 | 0.16 |
ENST00000605176.1 |
NBPF10 |
neuroblastoma breakpoint family, member 10 |
| chr17_-_8263538 | 0.15 |
ENST00000535173.1 |
AC135178.1 |
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr5_-_90679145 | 0.15 |
ENST00000265138.3 |
ARRDC3 |
arrestin domain containing 3 |
| chr11_+_131240373 | 0.15 |
ENST00000374791.3 ENST00000436745.1 |
NTM |
neurotrimin |
| chr1_+_145293371 | 0.15 |
ENST00000342960.5 |
NBPF10 |
neuroblastoma breakpoint family, member 10 |
| chr2_-_55646412 | 0.14 |
ENST00000413716.2 |
CCDC88A |
coiled-coil domain containing 88A |
| chr19_-_58864848 | 0.14 |
ENST00000263100.3 |
A1BG |
alpha-1-B glycoprotein |
| chr3_+_152017360 | 0.14 |
ENST00000485910.1 ENST00000463374.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr12_-_90024360 | 0.14 |
ENST00000393164.2 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr11_+_30253410 | 0.14 |
ENST00000533718.1 |
FSHB |
follicle stimulating hormone, beta polypeptide |
| chr1_+_113933581 | 0.13 |
ENST00000307546.9 ENST00000369615.1 ENST00000369611.4 |
MAGI3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr7_-_132261253 | 0.13 |
ENST00000321063.4 |
PLXNA4 |
plexin A4 |
| chr5_-_171881362 | 0.13 |
ENST00000519643.1 |
SH3PXD2B |
SH3 and PX domains 2B |
| chr10_-_53459319 | 0.13 |
ENST00000331173.4 |
CSTF2T |
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
| chr3_+_101546827 | 0.13 |
ENST00000461724.1 ENST00000483180.1 ENST00000394054.2 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr10_-_28270795 | 0.13 |
ENST00000545014.1 |
ARMC4 |
armadillo repeat containing 4 |
| chr10_+_63661053 | 0.13 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr12_+_15475462 | 0.13 |
ENST00000543886.1 ENST00000348962.2 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr11_+_102980126 | 0.12 |
ENST00000375735.2 |
DYNC2H1 |
dynein, cytoplasmic 2, heavy chain 1 |
| chr19_-_19729725 | 0.12 |
ENST00000251203.9 |
PBX4 |
pre-B-cell leukemia homeobox 4 |
| chr7_+_142985308 | 0.12 |
ENST00000310447.5 |
CASP2 |
caspase 2, apoptosis-related cysteine peptidase |
| chr11_-_19223523 | 0.12 |
ENST00000265968.3 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr11_+_134123389 | 0.11 |
ENST00000281182.4 ENST00000537423.1 ENST00000543332.1 ENST00000374752.4 |
ACAD8 |
acyl-CoA dehydrogenase family, member 8 |
| chr8_-_135522425 | 0.11 |
ENST00000521673.1 |
ZFAT |
zinc finger and AT hook domain containing |
| chr11_+_102980251 | 0.11 |
ENST00000334267.7 ENST00000398093.3 |
DYNC2H1 |
dynein, cytoplasmic 2, heavy chain 1 |
| chr3_+_152017181 | 0.11 |
ENST00000498502.1 ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr5_-_171881491 | 0.11 |
ENST00000311601.5 |
SH3PXD2B |
SH3 and PX domains 2B |
| chr12_-_10978957 | 0.11 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr7_-_121784285 | 0.10 |
ENST00000417368.2 |
AASS |
aminoadipate-semialdehyde synthase |
| chr2_-_161350305 | 0.10 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr1_+_207262578 | 0.10 |
ENST00000243611.5 ENST00000367076.3 |
C4BPB |
complement component 4 binding protein, beta |
| chr6_+_27925019 | 0.10 |
ENST00000244623.1 |
OR2B6 |
olfactory receptor, family 2, subfamily B, member 6 |
| chr15_-_99789736 | 0.10 |
ENST00000560235.1 ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23 |
tetratricopeptide repeat domain 23 |
| chr17_-_26697304 | 0.10 |
ENST00000536498.1 |
VTN |
vitronectin |
| chr2_+_157330081 | 0.10 |
ENST00000409674.1 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr5_+_59726565 | 0.10 |
ENST00000412930.2 |
FKSG52 |
FKSG52 |
| chr19_-_45873642 | 0.09 |
ENST00000485403.2 ENST00000586856.1 ENST00000586131.1 ENST00000391940.4 ENST00000221481.6 ENST00000391944.3 ENST00000391945.4 |
ERCC2 |
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr17_-_79876010 | 0.09 |
ENST00000328666.6 |
SIRT7 |
sirtuin 7 |
| chr12_-_95941987 | 0.09 |
ENST00000537435.2 |
USP44 |
ubiquitin specific peptidase 44 |
| chr1_-_165668100 | 0.09 |
ENST00000354775.4 |
ALDH9A1 |
aldehyde dehydrogenase 9 family, member A1 |
| chr16_-_20702578 | 0.09 |
ENST00000307493.4 ENST00000219151.4 |
ACSM1 |
acyl-CoA synthetase medium-chain family member 1 |
| chr19_+_13135386 | 0.09 |
ENST00000360105.4 ENST00000588228.1 ENST00000591028.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr3_-_164914640 | 0.09 |
ENST00000241274.3 |
SLITRK3 |
SLIT and NTRK-like family, member 3 |
| chr1_+_78245303 | 0.09 |
ENST00000370791.3 ENST00000443751.2 |
FAM73A |
family with sequence similarity 73, member A |
| chr4_+_187187098 | 0.09 |
ENST00000403665.2 ENST00000264692.4 |
F11 |
coagulation factor XI |
| chr4_+_123073464 | 0.09 |
ENST00000264501.4 |
KIAA1109 |
KIAA1109 |
| chr15_-_64995399 | 0.09 |
ENST00000559753.1 ENST00000560258.2 ENST00000559912.2 ENST00000326005.6 |
OAZ2 |
ornithine decarboxylase antizyme 2 |
| chr9_-_116840728 | 0.09 |
ENST00000265132.3 |
AMBP |
alpha-1-microglobulin/bikunin precursor |
| chr3_+_108308513 | 0.08 |
ENST00000361582.3 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
| chr10_-_127505167 | 0.08 |
ENST00000368786.1 |
UROS |
uroporphyrinogen III synthase |
| chr9_-_77703115 | 0.08 |
ENST00000361092.4 ENST00000376808.4 |
NMRK1 |
nicotinamide riboside kinase 1 |
| chrX_+_135388147 | 0.08 |
ENST00000394141.1 |
GPR112 |
G protein-coupled receptor 112 |
| chr1_+_207262627 | 0.08 |
ENST00000391923.1 |
C4BPB |
complement component 4 binding protein, beta |
| chr1_+_156756667 | 0.08 |
ENST00000526188.1 ENST00000454659.1 |
PRCC |
papillary renal cell carcinoma (translocation-associated) |
| chr13_+_30002846 | 0.08 |
ENST00000542829.1 |
MTUS2 |
microtubule associated tumor suppressor candidate 2 |
| chrM_-_14670 | 0.08 |
ENST00000361681.2 |
MT-ND6 |
mitochondrially encoded NADH dehydrogenase 6 |
| chrX_-_71792477 | 0.08 |
ENST00000421523.1 ENST00000415409.1 ENST00000373559.4 ENST00000373556.3 ENST00000373560.2 ENST00000373583.1 ENST00000429103.2 ENST00000373571.1 ENST00000373554.1 |
HDAC8 |
histone deacetylase 8 |
| chr15_+_31196053 | 0.08 |
ENST00000561594.1 ENST00000362065.4 |
FAN1 |
FANCD2/FANCI-associated nuclease 1 |
| chr8_-_145028013 | 0.08 |
ENST00000354958.2 |
PLEC |
plectin |
| chrX_-_16887963 | 0.08 |
ENST00000380084.4 |
RBBP7 |
retinoblastoma binding protein 7 |
| chr1_+_207262540 | 0.08 |
ENST00000452902.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr12_-_110434183 | 0.08 |
ENST00000360185.4 ENST00000354574.4 ENST00000338373.5 ENST00000343646.5 ENST00000356259.4 ENST00000553118.1 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr4_+_159727272 | 0.08 |
ENST00000379346.3 |
FNIP2 |
folliculin interacting protein 2 |
| chr12_-_108954933 | 0.07 |
ENST00000431469.2 ENST00000546815.1 |
SART3 |
squamous cell carcinoma antigen recognized by T cells 3 |
| chr12_-_54653313 | 0.07 |
ENST00000550411.1 ENST00000439541.2 |
CBX5 |
chromobox homolog 5 |
| chr3_-_52273098 | 0.07 |
ENST00000499914.2 ENST00000305533.5 ENST00000597542.1 |
TWF2 TLR9 |
twinfilin actin-binding protein 2 toll-like receptor 9 |
| chr17_-_56082455 | 0.07 |
ENST00000578794.1 |
RP11-159D12.5 |
Uncharacterized protein |
| chr10_-_46167722 | 0.07 |
ENST00000374366.3 ENST00000344646.5 |
ZFAND4 |
zinc finger, AN1-type domain 4 |
| chr14_+_74083548 | 0.07 |
ENST00000381139.1 |
ACOT6 |
acyl-CoA thioesterase 6 |
| chr12_-_110434021 | 0.07 |
ENST00000355312.3 ENST00000551209.1 ENST00000550186.1 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr14_+_57671888 | 0.07 |
ENST00000391612.1 |
AL391152.1 |
AL391152.1 |
| chr20_+_36373032 | 0.07 |
ENST00000373473.1 |
CTNNBL1 |
catenin, beta like 1 |
| chr11_+_118938485 | 0.07 |
ENST00000300793.6 |
VPS11 |
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chr15_+_31196080 | 0.06 |
ENST00000561607.1 ENST00000565466.1 |
FAN1 |
FANCD2/FANCI-associated nuclease 1 |
| chr9_-_14180778 | 0.06 |
ENST00000380924.1 ENST00000543693.1 |
NFIB |
nuclear factor I/B |
| chr1_+_159557607 | 0.06 |
ENST00000255040.2 |
APCS |
amyloid P component, serum |
| chr5_+_42756903 | 0.06 |
ENST00000361970.5 ENST00000388827.4 |
CCDC152 |
coiled-coil domain containing 152 |
| chr1_+_207262881 | 0.06 |
ENST00000451804.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr4_+_169418255 | 0.06 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr22_-_36220420 | 0.06 |
ENST00000473487.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr20_+_22034809 | 0.06 |
ENST00000449427.1 |
RP11-125P18.1 |
RP11-125P18.1 |
| chr6_+_127898312 | 0.05 |
ENST00000329722.7 |
C6orf58 |
chromosome 6 open reading frame 58 |
| chr16_-_75467318 | 0.05 |
ENST00000283882.3 |
CFDP1 |
craniofacial development protein 1 |
| chr14_+_24422795 | 0.05 |
ENST00000313250.5 ENST00000558263.1 ENST00000543741.2 ENST00000421831.1 ENST00000397073.2 ENST00000308178.8 ENST00000382761.3 ENST00000397075.3 ENST00000397074.3 ENST00000559632.1 ENST00000558581.1 |
DHRS4 |
dehydrogenase/reductase (SDR family) member 4 |
| chr5_+_169780485 | 0.05 |
ENST00000377360.4 |
KCNIP1 |
Kv channel interacting protein 1 |
| chr4_+_37245799 | 0.05 |
ENST00000309447.5 |
KIAA1239 |
KIAA1239 |
| chr22_+_51176624 | 0.05 |
ENST00000216139.5 ENST00000529621.1 |
ACR |
acrosin |
| chrX_+_78426469 | 0.05 |
ENST00000276077.1 |
GPR174 |
G protein-coupled receptor 174 |
| chr9_+_80850952 | 0.05 |
ENST00000424347.2 ENST00000415759.2 ENST00000376597.4 ENST00000277082.5 ENST00000376598.2 |
CEP78 |
centrosomal protein 78kDa |
| chr17_-_50235423 | 0.05 |
ENST00000340813.6 |
CA10 |
carbonic anhydrase X |
| chr8_-_70983506 | 0.05 |
ENST00000276594.2 |
PRDM14 |
PR domain containing 14 |
| chr9_-_77703056 | 0.05 |
ENST00000376811.1 |
NMRK1 |
nicotinamide riboside kinase 1 |
| chr11_+_105948216 | 0.05 |
ENST00000278618.4 |
AASDHPPT |
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr21_+_17961006 | 0.05 |
ENST00000602323.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr13_-_46679144 | 0.05 |
ENST00000181383.4 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr3_+_127770455 | 0.05 |
ENST00000464451.1 |
SEC61A1 |
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr15_+_38746307 | 0.04 |
ENST00000397609.2 ENST00000491535.1 |
FAM98B |
family with sequence similarity 98, member B |
| chr18_+_49866496 | 0.04 |
ENST00000442544.2 |
DCC |
deleted in colorectal carcinoma |
| chr4_+_169418195 | 0.04 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr18_+_11490077 | 0.04 |
ENST00000586947.1 ENST00000591431.1 |
RP11-712C7.1 |
RP11-712C7.1 |
| chr15_-_47426320 | 0.04 |
ENST00000557832.1 |
FKSG62 |
FKSG62 |
| chr13_-_107187462 | 0.04 |
ENST00000245323.4 |
EFNB2 |
ephrin-B2 |
| chr1_-_159684371 | 0.04 |
ENST00000255030.5 ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP |
C-reactive protein, pentraxin-related |
| chr2_-_161349909 | 0.04 |
ENST00000392753.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr1_+_153232160 | 0.04 |
ENST00000368742.3 |
LOR |
loricrin |
| chr13_-_46679185 | 0.04 |
ENST00000439329.3 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chrX_+_54466829 | 0.04 |
ENST00000375151.4 |
TSR2 |
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr14_+_37131058 | 0.04 |
ENST00000361487.6 |
PAX9 |
paired box 9 |
| chr2_-_203735586 | 0.04 |
ENST00000454326.1 ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr17_+_56232494 | 0.04 |
ENST00000268912.5 |
OR4D1 |
olfactory receptor, family 4, subfamily D, member 1 |
| chr14_-_21056121 | 0.04 |
ENST00000557105.1 ENST00000398008.2 ENST00000555841.1 ENST00000443456.2 ENST00000432835.2 ENST00000557503.1 ENST00000398009.2 ENST00000554842.1 |
RNASE11 |
ribonuclease, RNase A family, 11 (non-active) |
| chr1_-_32384693 | 0.04 |
ENST00000602683.1 ENST00000470404.1 |
PTP4A2 |
protein tyrosine phosphatase type IVA, member 2 |
| chr21_-_43816052 | 0.04 |
ENST00000398405.1 |
TMPRSS3 |
transmembrane protease, serine 3 |
| chr10_+_51576285 | 0.04 |
ENST00000443446.1 |
NCOA4 |
nuclear receptor coactivator 4 |
| chr11_-_105948040 | 0.04 |
ENST00000534815.1 |
KBTBD3 |
kelch repeat and BTB (POZ) domain containing 3 |
| chr4_+_96761238 | 0.03 |
ENST00000295266.4 |
PDHA2 |
pyruvate dehydrogenase (lipoamide) alpha 2 |
| chr6_-_46048116 | 0.03 |
ENST00000185206.6 |
CLIC5 |
chloride intracellular channel 5 |
| chr2_-_85788652 | 0.03 |
ENST00000430215.3 |
GGCX |
gamma-glutamyl carboxylase |
| chr12_+_15475331 | 0.03 |
ENST00000281171.4 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr1_-_1655713 | 0.03 |
ENST00000401096.2 ENST00000404249.3 ENST00000357760.2 ENST00000358779.5 ENST00000378633.1 ENST00000378635.3 |
CDK11A |
cyclin-dependent kinase 11A |
| chr2_+_234959376 | 0.03 |
ENST00000425558.1 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
| chr19_+_45596398 | 0.03 |
ENST00000544069.2 |
PPP1R37 |
protein phosphatase 1, regulatory subunit 37 |
| chr11_-_95657231 | 0.03 |
ENST00000409459.1 ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2 |
myotubularin related protein 2 |
| chr16_+_69958887 | 0.03 |
ENST00000568684.1 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
| chr10_-_35104185 | 0.03 |
ENST00000374789.3 ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3 |
par-3 family cell polarity regulator |
| chr10_+_98064085 | 0.03 |
ENST00000419175.1 ENST00000371174.2 |
DNTT |
DNA nucleotidylexotransferase |
| chr5_+_180794269 | 0.03 |
ENST00000456475.1 |
OR4F3 |
olfactory receptor, family 4, subfamily F, member 3 |
| chr4_-_168155169 | 0.03 |
ENST00000534949.1 ENST00000535728.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr10_+_115312766 | 0.03 |
ENST00000351270.3 |
HABP2 |
hyaluronan binding protein 2 |
| chr12_+_111471828 | 0.03 |
ENST00000261726.6 |
CUX2 |
cut-like homeobox 2 |
| chrX_+_122318113 | 0.03 |
ENST00000371264.3 |
GRIA3 |
glutamate receptor, ionotropic, AMPA 3 |
| chr11_-_10828892 | 0.03 |
ENST00000525681.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr8_+_121457642 | 0.03 |
ENST00000305949.1 |
MTBP |
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
| chr14_-_21055874 | 0.03 |
ENST00000553849.1 |
RNASE11 |
ribonuclease, RNase A family, 11 (non-active) |
| chr12_-_13529642 | 0.03 |
ENST00000318426.2 |
C12orf36 |
chromosome 12 open reading frame 36 |
| chr19_+_11200038 | 0.03 |
ENST00000558518.1 ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR |
low density lipoprotein receptor |
| chr18_-_31802056 | 0.03 |
ENST00000538587.1 |
NOL4 |
nucleolar protein 4 |
| chr2_+_149804382 | 0.03 |
ENST00000397413.1 |
KIF5C |
kinesin family member 5C |
| chr20_+_21686290 | 0.02 |
ENST00000398485.2 |
PAX1 |
paired box 1 |
| chr21_-_27107283 | 0.02 |
ENST00000284971.3 ENST00000400099.1 |
ATP5J |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr2_+_108602971 | 0.02 |
ENST00000409059.1 ENST00000540517.1 ENST00000264047.2 |
SLC5A7 |
solute carrier family 5 (sodium/choline cotransporter), member 7 |
| chr17_+_39868577 | 0.02 |
ENST00000329402.3 |
GAST |
gastrin |
| chr18_-_31802282 | 0.02 |
ENST00000535475.1 |
NOL4 |
nucleolar protein 4 |
| chr4_+_76649797 | 0.02 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr3_+_186383741 | 0.02 |
ENST00000232003.4 |
HRG |
histidine-rich glycoprotein |
| chr1_-_162346657 | 0.02 |
ENST00000367935.5 |
C1orf111 |
chromosome 1 open reading frame 111 |
| chr7_-_32529973 | 0.02 |
ENST00000410044.1 ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5 |
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr11_-_105948129 | 0.02 |
ENST00000526793.1 |
KBTBD3 |
kelch repeat and BTB (POZ) domain containing 3 |
| chr13_+_111267866 | 0.02 |
ENST00000458711.2 ENST00000424185.2 ENST00000397191.4 ENST00000309957.2 |
CARKD |
carbohydrate kinase domain containing |
| chr11_-_122931881 | 0.02 |
ENST00000526110.1 ENST00000227378.3 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr5_+_128300810 | 0.02 |
ENST00000262462.4 |
SLC27A6 |
solute carrier family 27 (fatty acid transporter), member 6 |
| chr21_-_43735628 | 0.02 |
ENST00000291525.10 ENST00000518498.1 |
TFF3 |
trefoil factor 3 (intestinal) |
| chr16_+_31044413 | 0.02 |
ENST00000394998.1 |
STX4 |
syntaxin 4 |
| chr1_-_94586651 | 0.02 |
ENST00000535735.1 ENST00000370225.3 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr1_-_26633067 | 0.02 |
ENST00000421827.2 ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11 |
UBX domain protein 11 |
| chr1_-_197036364 | 0.02 |
ENST00000367412.1 |
F13B |
coagulation factor XIII, B polypeptide |
| chr2_-_197457335 | 0.02 |
ENST00000260983.3 |
HECW2 |
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr21_-_27107344 | 0.02 |
ENST00000457143.2 |
ATP5J |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr18_-_24765248 | 0.02 |
ENST00000580774.1 ENST00000284224.8 |
CHST9 |
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr1_+_57320437 | 0.02 |
ENST00000361249.3 |
C8A |
complement component 8, alpha polypeptide |
| chr21_-_43816152 | 0.02 |
ENST00000433957.2 ENST00000398397.3 |
TMPRSS3 |
transmembrane protease, serine 3 |
| chr13_-_36920420 | 0.02 |
ENST00000438666.2 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr2_-_214016314 | 0.02 |
ENST00000434687.1 ENST00000374319.4 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
| chr19_+_45449301 | 0.02 |
ENST00000591597.1 |
APOC2 |
apolipoprotein C-II |
| chr8_-_121457608 | 0.02 |
ENST00000306185.3 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr21_-_27107198 | 0.02 |
ENST00000400094.1 |
ATP5J |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr4_-_103746683 | 0.01 |
ENST00000504211.1 ENST00000508476.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr6_+_2246023 | 0.01 |
ENST00000530833.1 ENST00000525811.1 ENST00000534441.1 ENST00000533653.1 ENST00000534468.1 |
GMDS-AS1 |
GMDS antisense RNA 1 (head to head) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.1 | 0.2 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.2 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 1.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.5 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.3 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.2 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) positive regulation of interferon-alpha biosynthetic process(GO:0045356) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.0 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.3 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |