Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
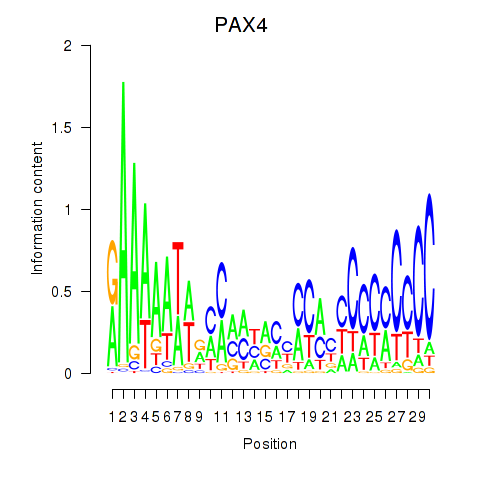
Results for PAX4
Z-value: 0.10
Transcription factors associated with PAX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX4
|
ENSG00000106331.10 | PAX4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX4 | hg19_v2_chr7_-_127255724_127255780 | 0.08 | 8.6e-01 | Click! |
Activity profile of PAX4 motif
Sorted Z-values of PAX4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_101702417 | 0.21 |
ENST00000305352.6 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
| chr8_+_17434689 | 0.18 |
ENST00000398074.3 |
PDGFRL |
platelet-derived growth factor receptor-like |
| chr7_+_80267973 | 0.17 |
ENST00000394788.3 ENST00000447544.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr9_-_20622478 | 0.16 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chrX_+_66764375 | 0.16 |
ENST00000374690.3 |
AR |
androgen receptor |
| chr6_+_32407619 | 0.12 |
ENST00000395388.2 ENST00000374982.5 |
HLA-DRA |
major histocompatibility complex, class II, DR alpha |
| chr4_+_39184024 | 0.12 |
ENST00000399820.3 ENST00000509560.1 ENST00000512112.1 ENST00000288634.7 ENST00000506503.1 |
WDR19 |
WD repeat domain 19 |
| chr17_+_47448102 | 0.11 |
ENST00000576461.1 |
RP11-81K2.1 |
Uncharacterized protein |
| chr21_+_26934165 | 0.11 |
ENST00000456917.1 |
MIR155HG |
MIR155 host gene (non-protein coding) |
| chr11_+_1874200 | 0.11 |
ENST00000311604.3 |
LSP1 |
lymphocyte-specific protein 1 |
| chr10_+_120116527 | 0.10 |
ENST00000445161.1 |
LINC00867 |
long intergenic non-protein coding RNA 867 |
| chr12_-_48164812 | 0.10 |
ENST00000549151.1 ENST00000548919.1 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr10_-_70092671 | 0.09 |
ENST00000358769.2 ENST00000432941.1 ENST00000495025.2 |
PBLD |
phenazine biosynthesis-like protein domain containing |
| chr15_+_69373184 | 0.09 |
ENST00000558147.1 ENST00000440444.1 |
LINC00277 |
long intergenic non-protein coding RNA 277 |
| chr19_+_2977444 | 0.09 |
ENST00000246112.4 ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6 |
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr5_-_20575959 | 0.09 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr17_+_1674982 | 0.09 |
ENST00000572048.1 ENST00000573763.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr16_-_49890016 | 0.09 |
ENST00000563137.2 |
ZNF423 |
zinc finger protein 423 |
| chr12_-_45269430 | 0.08 |
ENST00000395487.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr19_-_51336443 | 0.08 |
ENST00000598673.1 |
KLK15 |
kallikrein-related peptidase 15 |
| chr2_+_185463093 | 0.08 |
ENST00000302277.6 |
ZNF804A |
zinc finger protein 804A |
| chr8_-_13134045 | 0.08 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr1_-_72748417 | 0.08 |
ENST00000357731.5 |
NEGR1 |
neuronal growth regulator 1 |
| chr7_-_99756293 | 0.08 |
ENST00000316937.3 ENST00000456769.1 |
C7orf43 |
chromosome 7 open reading frame 43 |
| chr19_-_36391434 | 0.08 |
ENST00000396901.1 ENST00000585925.1 |
NFKBID |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta |
| chr9_-_5185629 | 0.08 |
ENST00000381641.3 |
INSL6 |
insulin-like 6 |
| chr13_+_76334498 | 0.08 |
ENST00000534657.1 |
LMO7 |
LIM domain 7 |
| chr4_+_47487285 | 0.07 |
ENST00000273859.3 ENST00000504445.1 |
ATP10D |
ATPase, class V, type 10D |
| chr8_-_116681221 | 0.07 |
ENST00000395715.3 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr16_+_53088885 | 0.07 |
ENST00000566029.1 ENST00000447540.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr14_+_104394770 | 0.07 |
ENST00000409874.4 ENST00000339063.5 |
TDRD9 |
tudor domain containing 9 |
| chr17_-_46799872 | 0.07 |
ENST00000290294.3 |
PRAC1 |
prostate cancer susceptibility candidate 1 |
| chr17_+_73089382 | 0.07 |
ENST00000538213.2 ENST00000584118.1 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr1_+_34326076 | 0.07 |
ENST00000519684.1 ENST00000522796.1 |
HMGB4 |
high mobility group box 4 |
| chr20_-_18810797 | 0.07 |
ENST00000278779.4 |
C20orf78 |
chromosome 20 open reading frame 78 |
| chr3_+_69134124 | 0.07 |
ENST00000478935.1 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr1_+_22889953 | 0.07 |
ENST00000374644.4 ENST00000166244.3 ENST00000538803.1 |
EPHA8 |
EPH receptor A8 |
| chr12_-_13248598 | 0.07 |
ENST00000337630.6 ENST00000545699.1 |
GSG1 |
germ cell associated 1 |
| chr1_+_15480197 | 0.06 |
ENST00000400796.3 ENST00000434578.2 ENST00000376008.2 |
TMEM51 |
transmembrane protein 51 |
| chr11_-_114466471 | 0.06 |
ENST00000424261.2 |
NXPE4 |
neurexophilin and PC-esterase domain family, member 4 |
| chr4_+_14113592 | 0.06 |
ENST00000502759.1 ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085 |
long intergenic non-protein coding RNA 1085 |
| chr10_-_25012115 | 0.06 |
ENST00000446003.1 |
ARHGAP21 |
Rho GTPase activating protein 21 |
| chr8_+_143781883 | 0.06 |
ENST00000522591.1 |
LY6K |
lymphocyte antigen 6 complex, locus K |
| chr2_+_102972363 | 0.06 |
ENST00000409599.1 |
IL18R1 |
interleukin 18 receptor 1 |
| chr8_+_21946681 | 0.06 |
ENST00000289921.7 |
FAM160B2 |
family with sequence similarity 160, member B2 |
| chr6_+_32605195 | 0.06 |
ENST00000374949.2 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr8_+_67344710 | 0.06 |
ENST00000379385.4 ENST00000396623.3 ENST00000415254.1 |
ADHFE1 |
alcohol dehydrogenase, iron containing, 1 |
| chr3_-_79816965 | 0.06 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr8_-_38853990 | 0.06 |
ENST00000456845.2 ENST00000397070.2 ENST00000517872.1 ENST00000412303.1 ENST00000456397.2 |
TM2D2 |
TM2 domain containing 2 |
| chrX_+_107683096 | 0.06 |
ENST00000328300.6 ENST00000361603.2 |
COL4A5 |
collagen, type IV, alpha 5 |
| chr3_-_9921934 | 0.06 |
ENST00000423850.1 |
CIDEC |
cell death-inducing DFFA-like effector c |
| chrX_+_65384052 | 0.06 |
ENST00000336279.5 ENST00000458621.1 |
HEPH |
hephaestin |
| chrX_+_65384182 | 0.06 |
ENST00000441993.2 ENST00000419594.1 |
HEPH |
hephaestin |
| chr1_-_231376836 | 0.06 |
ENST00000451322.1 |
C1orf131 |
chromosome 1 open reading frame 131 |
| chr11_+_22689648 | 0.06 |
ENST00000278187.3 |
GAS2 |
growth arrest-specific 2 |
| chrX_+_135067576 | 0.06 |
ENST00000370701.1 ENST00000370698.3 ENST00000370695.4 |
SLC9A6 |
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6 |
| chr1_-_1624083 | 0.06 |
ENST00000378662.1 ENST00000234800.6 |
SLC35E2B |
solute carrier family 35, member E2B |
| chr8_-_116680833 | 0.06 |
ENST00000220888.5 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr2_+_203879568 | 0.05 |
ENST00000449802.1 |
NBEAL1 |
neurobeachin-like 1 |
| chr19_+_49891475 | 0.05 |
ENST00000447857.3 |
CCDC155 |
coiled-coil domain containing 155 |
| chr3_+_186383741 | 0.05 |
ENST00000232003.4 |
HRG |
histidine-rich glycoprotein |
| chr8_+_143781513 | 0.05 |
ENST00000292430.6 ENST00000561179.1 ENST00000518841.1 ENST00000519387.1 |
LY6K |
lymphocyte antigen 6 complex, locus K |
| chr4_-_171011323 | 0.05 |
ENST00000509167.1 ENST00000353187.2 ENST00000507375.1 ENST00000515480.1 |
AADAT |
aminoadipate aminotransferase |
| chrX_-_107682702 | 0.05 |
ENST00000372216.4 |
COL4A6 |
collagen, type IV, alpha 6 |
| chr11_-_114466477 | 0.05 |
ENST00000375478.3 |
NXPE4 |
neurexophilin and PC-esterase domain family, member 4 |
| chr6_+_31554636 | 0.05 |
ENST00000433492.1 |
LST1 |
leukocyte specific transcript 1 |
| chr3_+_130569429 | 0.05 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr1_+_155146318 | 0.05 |
ENST00000368385.4 ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46 |
tripartite motif containing 46 |
| chr2_-_85555385 | 0.05 |
ENST00000377386.3 |
TGOLN2 |
trans-golgi network protein 2 |
| chr1_+_210001309 | 0.05 |
ENST00000491415.2 |
DIEXF |
digestive organ expansion factor homolog (zebrafish) |
| chr13_+_37247934 | 0.05 |
ENST00000315190.3 |
SERTM1 |
serine-rich and transmembrane domain containing 1 |
| chr7_-_140098257 | 0.05 |
ENST00000340308.3 ENST00000447932.2 ENST00000429996.2 ENST00000469193.1 ENST00000326232.9 |
SLC37A3 |
solute carrier family 37, member 3 |
| chr11_-_111741994 | 0.05 |
ENST00000398006.2 |
ALG9 |
ALG9, alpha-1,2-mannosyltransferase |
| chr1_-_155145721 | 0.05 |
ENST00000295682.4 |
KRTCAP2 |
keratinocyte associated protein 2 |
| chr13_+_76334795 | 0.05 |
ENST00000526202.1 ENST00000465261.2 |
LMO7 |
LIM domain 7 |
| chr10_-_70092635 | 0.05 |
ENST00000309049.4 |
PBLD |
phenazine biosynthesis-like protein domain containing |
| chr7_-_130080977 | 0.05 |
ENST00000223208.5 |
CEP41 |
centrosomal protein 41kDa |
| chrX_+_110187513 | 0.04 |
ENST00000446737.1 ENST00000425146.1 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr15_+_63340647 | 0.04 |
ENST00000404484.4 |
TPM1 |
tropomyosin 1 (alpha) |
| chr7_+_66386204 | 0.04 |
ENST00000341567.4 ENST00000607045.1 |
TMEM248 |
transmembrane protein 248 |
| chr12_+_122181529 | 0.04 |
ENST00000541467.1 |
TMEM120B |
transmembrane protein 120B |
| chr5_-_58882219 | 0.04 |
ENST00000505453.1 ENST00000360047.5 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr1_+_15802594 | 0.04 |
ENST00000375910.3 |
CELA2B |
chymotrypsin-like elastase family, member 2B |
| chr11_+_72975524 | 0.04 |
ENST00000540342.1 ENST00000542092.1 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr1_-_231376867 | 0.04 |
ENST00000366649.2 ENST00000318906.2 ENST00000366651.3 |
C1orf131 |
chromosome 1 open reading frame 131 |
| chr11_+_72975578 | 0.04 |
ENST00000393592.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr17_+_72322346 | 0.04 |
ENST00000551294.1 ENST00000389916.4 |
KIF19 |
kinesin family member 19 |
| chrM_+_5824 | 0.04 |
ENST00000361624.2 |
MT-CO1 |
mitochondrially encoded cytochrome c oxidase I |
| chr20_-_34117447 | 0.04 |
ENST00000246199.2 ENST00000424444.1 ENST00000374345.4 ENST00000444723.1 |
C20orf173 |
chromosome 20 open reading frame 173 |
| chr6_-_25042231 | 0.04 |
ENST00000510784.2 |
FAM65B |
family with sequence similarity 65, member B |
| chr11_+_72975559 | 0.04 |
ENST00000349767.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr4_+_492985 | 0.04 |
ENST00000296306.7 ENST00000536264.1 ENST00000310340.5 ENST00000453061.2 ENST00000504346.1 ENST00000503111.1 ENST00000383028.4 ENST00000509768.1 |
PIGG |
phosphatidylinositol glycan anchor biosynthesis, class G |
| chr21_-_21630994 | 0.04 |
ENST00000436373.1 |
AP001171.1 |
AP001171.1 |
| chr7_-_137686791 | 0.04 |
ENST00000452463.1 ENST00000330387.6 ENST00000456390.1 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
| chr1_+_33439268 | 0.04 |
ENST00000594612.1 |
FKSG48 |
FKSG48 |
| chrX_-_48827976 | 0.04 |
ENST00000218176.3 |
KCND1 |
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr1_-_1677358 | 0.04 |
ENST00000355439.2 ENST00000400924.1 ENST00000246421.4 |
SLC35E2 |
solute carrier family 35, member E2 |
| chr1_-_209957882 | 0.04 |
ENST00000294811.1 |
C1orf74 |
chromosome 1 open reading frame 74 |
| chr19_-_52551814 | 0.04 |
ENST00000594154.1 ENST00000598745.1 ENST00000597273.1 |
ZNF432 |
zinc finger protein 432 |
| chr2_+_217735493 | 0.04 |
ENST00000456163.1 |
AC007557.1 |
HCG1816075; Uncharacterized protein |
| chr13_+_76334567 | 0.04 |
ENST00000321797.8 |
LMO7 |
LIM domain 7 |
| chr9_-_100000957 | 0.03 |
ENST00000366109.2 ENST00000607322.1 |
RP11-498P14.5 |
RP11-498P14.5 |
| chr4_+_170541835 | 0.03 |
ENST00000504131.2 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr11_-_62474803 | 0.03 |
ENST00000533982.1 ENST00000360796.5 |
BSCL2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr1_-_151032040 | 0.03 |
ENST00000540998.1 ENST00000357235.5 |
CDC42SE1 |
CDC42 small effector 1 |
| chr3_-_197476560 | 0.03 |
ENST00000273582.5 |
KIAA0226 |
KIAA0226 |
| chr13_+_28712614 | 0.03 |
ENST00000380958.3 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr3_+_51895621 | 0.03 |
ENST00000333127.3 |
IQCF2 |
IQ motif containing F2 |
| chr4_+_170541678 | 0.03 |
ENST00000360642.3 ENST00000512813.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr12_+_31812121 | 0.03 |
ENST00000395763.3 |
METTL20 |
methyltransferase like 20 |
| chr16_-_67224002 | 0.03 |
ENST00000563889.1 ENST00000564418.1 ENST00000545725.2 ENST00000314586.6 |
EXOC3L1 |
exocyst complex component 3-like 1 |
| chr9_+_70971815 | 0.03 |
ENST00000396392.1 ENST00000396396.1 |
PGM5 |
phosphoglucomutase 5 |
| chr19_-_51961702 | 0.03 |
ENST00000430817.1 ENST00000321424.3 ENST00000340550.5 |
SIGLEC8 |
sialic acid binding Ig-like lectin 8 |
| chr19_-_37064145 | 0.03 |
ENST00000591340.1 ENST00000334116.7 |
ZNF529 |
zinc finger protein 529 |
| chrX_-_47004878 | 0.03 |
ENST00000377811.3 |
NDUFB11 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr2_-_136875712 | 0.03 |
ENST00000241393.3 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
| chr4_+_3465027 | 0.03 |
ENST00000389653.2 ENST00000507039.1 ENST00000340083.5 |
DOK7 |
docking protein 7 |
| chr6_-_32806506 | 0.03 |
ENST00000374897.2 ENST00000452392.2 |
TAP2 TAP2 |
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr2_-_217559517 | 0.03 |
ENST00000449583.1 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
| chr9_+_100000717 | 0.03 |
ENST00000375205.2 ENST00000357054.1 ENST00000395220.1 ENST00000375202.2 ENST00000411667.2 |
CCDC180 |
coiled-coil domain containing 180 |
| chr11_+_61891445 | 0.03 |
ENST00000394818.3 ENST00000533896.1 ENST00000278849.4 |
INCENP |
inner centromere protein antigens 135/155kDa |
| chr1_-_208084729 | 0.03 |
ENST00000310833.7 ENST00000356522.4 |
CD34 |
CD34 molecule |
| chr7_+_100482595 | 0.03 |
ENST00000448764.1 |
SRRT |
serrate RNA effector molecule homolog (Arabidopsis) |
| chr8_+_145215928 | 0.03 |
ENST00000528919.1 |
MROH1 |
maestro heat-like repeat family member 1 |
| chr2_-_170550877 | 0.03 |
ENST00000447353.1 |
CCDC173 |
coiled-coil domain containing 173 |
| chr7_+_133812052 | 0.03 |
ENST00000285928.2 |
LRGUK |
leucine-rich repeats and guanylate kinase domain containing |
| chr1_+_21880560 | 0.03 |
ENST00000425315.2 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
| chr4_+_170541660 | 0.03 |
ENST00000513761.1 ENST00000347613.4 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr1_-_151319283 | 0.03 |
ENST00000392746.3 |
RFX5 |
regulatory factor X, 5 (influences HLA class II expression) |
| chr5_-_172662303 | 0.03 |
ENST00000517440.1 ENST00000329198.4 |
NKX2-5 |
NK2 homeobox 5 |
| chr11_+_105481612 | 0.02 |
ENST00000531011.1 ENST00000525187.1 ENST00000530497.1 |
GRIA4 |
glutamate receptor, ionotropic, AMPA 4 |
| chr4_-_140544386 | 0.02 |
ENST00000561977.1 |
RP11-308D13.3 |
RP11-308D13.3 |
| chr4_+_165675269 | 0.02 |
ENST00000507311.1 |
RP11-294O2.2 |
RP11-294O2.2 |
| chr16_-_28608424 | 0.02 |
ENST00000335715.4 |
SULT1A2 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chrX_-_131623982 | 0.02 |
ENST00000370844.1 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr5_+_143584814 | 0.02 |
ENST00000507359.3 |
KCTD16 |
potassium channel tetramerization domain containing 16 |
| chr7_-_126892303 | 0.02 |
ENST00000358373.3 |
GRM8 |
glutamate receptor, metabotropic 8 |
| chr12_-_11091862 | 0.02 |
ENST00000537503.1 |
TAS2R14 |
taste receptor, type 2, member 14 |
| chr2_+_190526111 | 0.02 |
ENST00000607062.1 ENST00000260952.4 ENST00000425590.1 ENST00000607535.1 ENST00000420250.1 ENST00000606910.1 ENST00000607690.1 ENST00000607829.1 |
ASNSD1 |
asparagine synthetase domain containing 1 |
| chr12_-_95044309 | 0.02 |
ENST00000261226.4 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
| chr16_+_14980632 | 0.02 |
ENST00000565655.1 |
NOMO1 |
NODAL modulator 1 |
| chrX_-_131547625 | 0.02 |
ENST00000394311.2 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr15_-_40401062 | 0.02 |
ENST00000354670.4 ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF |
Bcl2 modifying factor |
| chr19_+_9361606 | 0.02 |
ENST00000456448.1 |
OR7E24 |
olfactory receptor, family 7, subfamily E, member 24 |
| chr1_+_26644441 | 0.02 |
ENST00000374213.2 |
CD52 |
CD52 molecule |
| chr4_+_30723003 | 0.02 |
ENST00000543491.1 |
PCDH7 |
protocadherin 7 |
| chr6_-_105627735 | 0.02 |
ENST00000254765.3 |
POPDC3 |
popeye domain containing 3 |
| chr17_-_77005801 | 0.02 |
ENST00000392446.5 |
CANT1 |
calcium activated nucleotidase 1 |
| chr18_-_53089723 | 0.02 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr2_-_85839146 | 0.02 |
ENST00000306336.5 ENST00000409734.3 |
C2orf68 |
chromosome 2 open reading frame 68 |
| chr4_+_75023816 | 0.02 |
ENST00000395759.2 ENST00000331145.6 ENST00000359107.5 ENST00000325278.6 |
MTHFD2L |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr11_+_120107344 | 0.02 |
ENST00000260264.4 |
POU2F3 |
POU class 2 homeobox 3 |
| chr12_-_113573495 | 0.02 |
ENST00000446861.3 |
RASAL1 |
RAS protein activator like 1 (GAP1 like) |
| chr11_+_46740730 | 0.02 |
ENST00000311907.5 ENST00000530231.1 ENST00000442468.1 |
F2 |
coagulation factor II (thrombin) |
| chr6_-_10694766 | 0.02 |
ENST00000460742.2 ENST00000259983.3 ENST00000379586.1 |
C6orf52 |
chromosome 6 open reading frame 52 |
| chr17_+_16284104 | 0.02 |
ENST00000577958.1 ENST00000302182.3 ENST00000577640.1 |
UBB |
ubiquitin B |
| chr3_+_39424828 | 0.02 |
ENST00000273158.4 ENST00000431510.1 |
SLC25A38 |
solute carrier family 25, member 38 |
| chr18_-_32957260 | 0.02 |
ENST00000587422.1 ENST00000306346.1 ENST00000589332.1 ENST00000586687.1 ENST00000585522.1 |
ZNF396 |
zinc finger protein 396 |
| chr19_+_55105085 | 0.02 |
ENST00000251372.3 ENST00000453777.1 |
LILRA1 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1 |
| chr5_+_146614579 | 0.02 |
ENST00000541094.1 ENST00000398521.3 |
STK32A |
serine/threonine kinase 32A |
| chr5_+_167913450 | 0.02 |
ENST00000231572.3 ENST00000538719.1 |
RARS |
arginyl-tRNA synthetase |
| chr3_-_71632894 | 0.02 |
ENST00000493089.1 |
FOXP1 |
forkhead box P1 |
| chrY_+_26997726 | 0.02 |
ENST00000382296.2 |
DAZ4 |
deleted in azoospermia 4 |
| chr17_+_43299241 | 0.02 |
ENST00000328118.3 |
FMNL1 |
formin-like 1 |
| chr12_-_51611477 | 0.02 |
ENST00000389243.4 |
POU6F1 |
POU class 6 homeobox 1 |
| chr3_+_111805182 | 0.02 |
ENST00000430855.1 ENST00000431717.2 ENST00000264848.5 |
C3orf52 |
chromosome 3 open reading frame 52 |
| chrX_-_131547596 | 0.02 |
ENST00000538204.1 ENST00000370849.3 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr17_-_77005860 | 0.02 |
ENST00000591773.1 ENST00000588611.1 ENST00000586916.2 ENST00000592033.1 ENST00000588075.1 ENST00000302345.2 ENST00000591811.1 |
CANT1 |
calcium activated nucleotidase 1 |
| chr5_-_133304473 | 0.02 |
ENST00000231512.3 |
C5orf15 |
chromosome 5 open reading frame 15 |
| chr1_+_152881014 | 0.02 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr1_+_168756151 | 0.02 |
ENST00000420691.1 |
LINC00626 |
long intergenic non-protein coding RNA 626 |
| chr11_-_62995986 | 0.02 |
ENST00000403374.2 |
SLC22A25 |
solute carrier family 22, member 25 |
| chr12_-_118406028 | 0.02 |
ENST00000425217.1 |
KSR2 |
kinase suppressor of ras 2 |
| chr4_+_109571800 | 0.02 |
ENST00000512478.2 |
OSTC |
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr7_-_130080818 | 0.02 |
ENST00000343969.5 ENST00000541543.1 ENST00000489512.1 |
CEP41 |
centrosomal protein 41kDa |
| chr8_+_104892639 | 0.02 |
ENST00000436393.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr18_-_53255766 | 0.02 |
ENST00000566286.1 ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4 |
transcription factor 4 |
| chr7_-_23053719 | 0.02 |
ENST00000432176.2 ENST00000440481.1 |
FAM126A |
family with sequence similarity 126, member A |
| chr4_+_95128996 | 0.02 |
ENST00000457823.2 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_95129061 | 0.02 |
ENST00000354268.4 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr22_-_31688431 | 0.02 |
ENST00000402249.3 ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chr14_+_76618242 | 0.02 |
ENST00000557542.1 ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L |
G patch domain containing 2-like |
| chr2_-_136873735 | 0.02 |
ENST00000409817.1 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
| chr5_-_142783175 | 0.02 |
ENST00000231509.3 ENST00000394464.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr5_-_126409159 | 0.02 |
ENST00000607731.1 ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63 |
chromosome 5 open reading frame 63 |
| chr19_-_51308175 | 0.02 |
ENST00000345523.4 |
C19orf48 |
chromosome 19 open reading frame 48 |
| chr6_-_32806483 | 0.01 |
ENST00000374899.4 |
TAP2 |
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr1_-_109618566 | 0.01 |
ENST00000338366.5 |
TAF13 |
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr12_-_120763739 | 0.01 |
ENST00000549767.1 |
PLA2G1B |
phospholipase A2, group IB (pancreas) |
| chr11_+_134144139 | 0.01 |
ENST00000389887.5 |
GLB1L3 |
galactosidase, beta 1-like 3 |
| chr1_+_231376941 | 0.01 |
ENST00000436239.1 ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT |
glyceronephosphate O-acyltransferase |
| chr6_-_32083106 | 0.01 |
ENST00000442721.1 |
TNXB |
tenascin XB |
| chr19_+_24216213 | 0.01 |
ENST00000594934.1 ENST00000597683.1 ENST00000342944.6 |
CTD-2017D11.1 ZNF254 |
CTD-2017D11.1 zinc finger protein 254 |
| chr5_-_142782862 | 0.01 |
ENST00000415690.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr17_+_4854375 | 0.01 |
ENST00000521811.1 ENST00000519602.1 ENST00000323997.6 ENST00000522249.1 ENST00000519584.1 |
ENO3 |
enolase 3 (beta, muscle) |
| chr1_+_21877753 | 0.01 |
ENST00000374832.1 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
| chr20_-_44993012 | 0.01 |
ENST00000372229.1 ENST00000372230.5 ENST00000543605.1 ENST00000243896.2 ENST00000317734.8 |
SLC35C2 |
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr17_+_16284399 | 0.01 |
ENST00000535788.1 |
UBB |
ubiquitin B |
| chr2_-_217560248 | 0.01 |
ENST00000233813.4 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
| chr12_-_57146095 | 0.01 |
ENST00000550770.1 ENST00000338193.6 |
PRIM1 |
primase, DNA, polypeptide 1 (49kDa) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 0.2 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.0 | GO:0072254 | metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) |
| 0.0 | 0.1 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.2 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.1 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.0 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.0 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.0 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |