Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for PAX7_NOBOX
Z-value: 1.10
Transcription factors associated with PAX7_NOBOX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
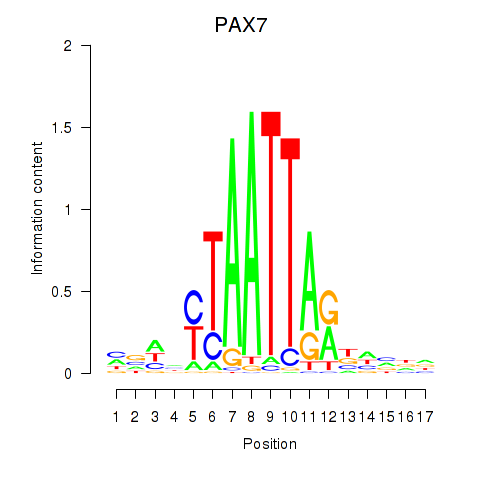
PAX7
|
ENSG00000009709.7 | PAX7 |
|
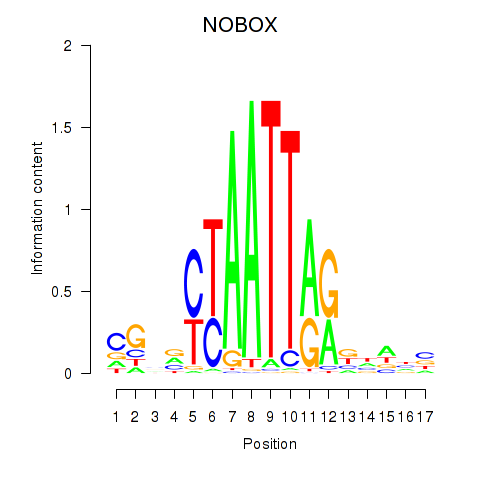
NOBOX
|
ENSG00000106410.10 | NOBOX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX7 | hg19_v2_chr1_+_18958008_18958023 | 0.53 | 1.8e-01 | Click! |
| NOBOX | hg19_v2_chr7_-_144100786_144100786 | 0.16 | 7.1e-01 | Click! |
Activity profile of PAX7_NOBOX motif
Sorted Z-values of PAX7_NOBOX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX7_NOBOX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_209602156 | 2.48 |
ENST00000429156.1 ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr19_-_36001113 | 1.50 |
ENST00000434389.1 |
DMKN |
dermokine |
| chr2_+_68961934 | 1.38 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr2_+_68961905 | 1.36 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr18_+_61254534 | 1.35 |
ENST00000269489.5 |
SERPINB13 |
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr10_-_105845674 | 1.30 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr1_+_28261492 | 1.18 |
ENST00000373894.3 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr18_+_61254570 | 1.09 |
ENST00000344731.5 |
SERPINB13 |
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr17_+_39394250 | 1.01 |
ENST00000254072.6 |
KRTAP9-8 |
keratin associated protein 9-8 |
| chr3_-_151034734 | 1.00 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr13_-_20806440 | 0.98 |
ENST00000400066.3 ENST00000400065.3 ENST00000356192.6 |
GJB6 |
gap junction protein, beta 6, 30kDa |
| chr1_+_62439037 | 0.98 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr17_-_19015945 | 0.85 |
ENST00000573866.2 |
SNORD3D |
small nucleolar RNA, C/D box 3D |
| chr1_+_160370344 | 0.83 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr11_+_117947724 | 0.82 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr2_+_68962014 | 0.81 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr4_-_74486217 | 0.80 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_+_66300446 | 0.79 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr13_-_86373536 | 0.78 |
ENST00000400286.2 |
SLITRK6 |
SLIT and NTRK-like family, member 6 |
| chr11_+_117947782 | 0.72 |
ENST00000522307.1 ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chrX_-_24690771 | 0.68 |
ENST00000379145.1 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr5_-_82969405 | 0.68 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr17_+_19091325 | 0.66 |
ENST00000584923.1 |
SNORD3A |
small nucleolar RNA, C/D box 3A |
| chr11_-_102668879 | 0.66 |
ENST00000315274.6 |
MMP1 |
matrix metallopeptidase 1 (interstitial collagenase) |
| chr12_+_107712173 | 0.64 |
ENST00000280758.5 ENST00000420571.2 |
BTBD11 |
BTB (POZ) domain containing 11 |
| chr17_+_39382900 | 0.61 |
ENST00000377721.3 ENST00000455970.2 |
KRTAP9-2 |
keratin associated protein 9-2 |
| chr11_-_119991589 | 0.57 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr1_+_81771806 | 0.56 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr12_+_26348246 | 0.54 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr8_+_105235572 | 0.53 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr6_+_47624172 | 0.52 |
ENST00000507065.1 ENST00000296862.1 |
GPR111 |
G protein-coupled receptor 111 |
| chr4_-_74486109 | 0.52 |
ENST00000395777.2 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr12_+_122688090 | 0.52 |
ENST00000324189.4 ENST00000546192.1 |
B3GNT4 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr1_+_186798073 | 0.50 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr6_+_26402465 | 0.49 |
ENST00000476549.2 ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chr20_-_50722183 | 0.49 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr17_-_7167279 | 0.46 |
ENST00000571932.2 |
CLDN7 |
claudin 7 |
| chr1_+_47533160 | 0.46 |
ENST00000334194.3 |
CYP4Z1 |
cytochrome P450, family 4, subfamily Z, polypeptide 1 |
| chr4_+_77356248 | 0.45 |
ENST00000296043.6 |
SHROOM3 |
shroom family member 3 |
| chr6_+_26365443 | 0.43 |
ENST00000527422.1 ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2 |
butyrophilin, subfamily 3, member A2 |
| chr3_-_141747950 | 0.43 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_+_111415757 | 0.41 |
ENST00000429072.2 ENST00000271324.5 |
CD53 |
CD53 molecule |
| chr19_+_11485333 | 0.41 |
ENST00000312423.2 |
SWSAP1 |
SWIM-type zinc finger 7 associated protein 1 |
| chr14_-_72458326 | 0.41 |
ENST00000542853.1 |
AC005477.1 |
AC005477.1 |
| chr11_-_7847519 | 0.40 |
ENST00000328375.1 |
OR5P3 |
olfactory receptor, family 5, subfamily P, member 3 |
| chr4_-_74486347 | 0.38 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr19_-_14945933 | 0.38 |
ENST00000322301.3 |
OR7A5 |
olfactory receptor, family 7, subfamily A, member 5 |
| chr5_+_140220769 | 0.37 |
ENST00000531613.1 ENST00000378123.3 |
PCDHA8 |
protocadherin alpha 8 |
| chr6_-_155776966 | 0.37 |
ENST00000159060.2 |
NOX3 |
NADPH oxidase 3 |
| chr9_+_124329336 | 0.36 |
ENST00000394340.3 ENST00000436835.1 ENST00000259371.2 |
DAB2IP |
DAB2 interacting protein |
| chr2_+_182850551 | 0.35 |
ENST00000452904.1 ENST00000409137.3 ENST00000280295.3 |
PPP1R1C |
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr1_+_233749739 | 0.35 |
ENST00000366621.3 |
KCNK1 |
potassium channel, subfamily K, member 1 |
| chr17_-_47841485 | 0.34 |
ENST00000506156.1 ENST00000240364.2 |
FAM117A |
family with sequence similarity 117, member A |
| chr5_-_95297534 | 0.32 |
ENST00000513343.1 ENST00000431061.2 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr18_+_44526786 | 0.32 |
ENST00000245121.5 ENST00000356157.7 |
KATNAL2 |
katanin p60 subunit A-like 2 |
| chr12_-_31479045 | 0.32 |
ENST00000539409.1 ENST00000395766.1 |
FAM60A |
family with sequence similarity 60, member A |
| chr16_-_28634874 | 0.32 |
ENST00000395609.1 ENST00000350842.4 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr11_-_124190184 | 0.31 |
ENST00000357438.2 |
OR8D2 |
olfactory receptor, family 8, subfamily D, member 2 |
| chr6_+_130339710 | 0.30 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr7_-_22234381 | 0.30 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr6_-_32806506 | 0.30 |
ENST00000374897.2 ENST00000452392.2 |
TAP2 TAP2 |
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr3_-_191000172 | 0.29 |
ENST00000427544.2 |
UTS2B |
urotensin 2B |
| chr8_-_109260897 | 0.29 |
ENST00000521297.1 ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E |
eukaryotic translation initiation factor 3, subunit E |
| chr6_+_26124373 | 0.28 |
ENST00000377791.2 ENST00000602637.1 |
HIST1H2AC |
histone cluster 1, H2ac |
| chr1_-_183560011 | 0.28 |
ENST00000367536.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr5_+_142149932 | 0.28 |
ENST00000274498.4 |
ARHGAP26 |
Rho GTPase activating protein 26 |
| chr8_-_91095099 | 0.28 |
ENST00000265431.3 |
CALB1 |
calbindin 1, 28kDa |
| chrX_+_138612889 | 0.28 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr11_-_59383617 | 0.28 |
ENST00000263847.1 |
OSBP |
oxysterol binding protein |
| chr5_-_35938674 | 0.28 |
ENST00000397366.1 ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL |
calcyphosine-like |
| chr1_-_183559693 | 0.28 |
ENST00000367535.3 ENST00000413720.1 ENST00000418089.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr14_+_22670455 | 0.28 |
ENST00000390460.1 |
TRAV26-2 |
T cell receptor alpha variable 26-2 |
| chr2_+_10091783 | 0.27 |
ENST00000324883.5 |
GRHL1 |
grainyhead-like 1 (Drosophila) |
| chr17_-_38821373 | 0.26 |
ENST00000394052.3 |
KRT222 |
keratin 222 |
| chr8_-_86290333 | 0.26 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr5_+_142149955 | 0.26 |
ENST00000378004.3 |
ARHGAP26 |
Rho GTPase activating protein 26 |
| chr14_-_104181771 | 0.26 |
ENST00000554913.1 ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3 |
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr20_+_31805131 | 0.26 |
ENST00000375454.3 ENST00000375452.3 |
BPIFA3 |
BPI fold containing family A, member 3 |
| chr4_-_25865159 | 0.25 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr12_+_26348429 | 0.25 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr4_+_86699834 | 0.25 |
ENST00000395183.2 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr9_-_95166841 | 0.25 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr2_+_102953608 | 0.24 |
ENST00000311734.2 ENST00000409584.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr4_-_89951028 | 0.24 |
ENST00000506913.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr10_+_24497704 | 0.23 |
ENST00000376456.4 ENST00000458595.1 |
KIAA1217 |
KIAA1217 |
| chr1_+_234509413 | 0.23 |
ENST00000366613.1 ENST00000366612.1 |
COA6 |
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr5_+_55147205 | 0.23 |
ENST00000396836.2 ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA |
interleukin 31 receptor A |
| chr15_-_51535208 | 0.23 |
ENST00000405913.3 ENST00000559878.1 |
CYP19A1 |
cytochrome P450, family 19, subfamily A, polypeptide 1 |
| chr9_+_139780942 | 0.23 |
ENST00000247668.2 ENST00000359662.3 |
TRAF2 |
TNF receptor-associated factor 2 |
| chr18_-_61329118 | 0.23 |
ENST00000332821.8 ENST00000283752.5 |
SERPINB3 |
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr2_-_101925055 | 0.23 |
ENST00000295317.3 |
RNF149 |
ring finger protein 149 |
| chr21_-_27423339 | 0.23 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr12_-_31479107 | 0.22 |
ENST00000542983.1 |
FAM60A |
family with sequence similarity 60, member A |
| chr15_+_58430567 | 0.22 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr12_+_66696322 | 0.22 |
ENST00000247815.4 |
HELB |
helicase (DNA) B |
| chr14_-_67878917 | 0.22 |
ENST00000216446.4 |
PLEK2 |
pleckstrin 2 |
| chr15_+_58430368 | 0.22 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr9_+_117904097 | 0.22 |
ENST00000374016.1 |
DEC1 |
deleted in esophageal cancer 1 |
| chr12_+_113354341 | 0.22 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr11_-_5323226 | 0.22 |
ENST00000380224.1 |
OR51B4 |
olfactory receptor, family 51, subfamily B, member 4 |
| chr4_-_46126093 | 0.22 |
ENST00000295452.4 |
GABRG1 |
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr17_-_62208169 | 0.21 |
ENST00000606895.1 |
ERN1 |
endoplasmic reticulum to nucleus signaling 1 |
| chr3_+_156009623 | 0.21 |
ENST00000389634.5 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr1_+_207226574 | 0.21 |
ENST00000367080.3 ENST00000367079.2 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr6_+_26440700 | 0.21 |
ENST00000494393.1 ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
| chr8_+_101170563 | 0.21 |
ENST00000520508.1 ENST00000388798.2 |
SPAG1 |
sperm associated antigen 1 |
| chr2_+_29320571 | 0.20 |
ENST00000401605.1 ENST00000401617.2 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
| chr18_+_616672 | 0.20 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr15_+_94899183 | 0.20 |
ENST00000557742.1 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
| chr12_-_31478428 | 0.19 |
ENST00000543615.1 |
FAM60A |
family with sequence similarity 60, member A |
| chr14_+_104182061 | 0.19 |
ENST00000216602.6 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr4_+_96012614 | 0.19 |
ENST00000264568.4 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr4_+_88896819 | 0.19 |
ENST00000237623.7 ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1 |
secreted phosphoprotein 1 |
| chr9_+_470288 | 0.19 |
ENST00000382303.1 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr10_-_99447024 | 0.18 |
ENST00000370626.3 |
AVPI1 |
arginine vasopressin-induced 1 |
| chr12_+_101988627 | 0.18 |
ENST00000547405.1 ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1 |
myosin binding protein C, slow type |
| chr12_+_101988774 | 0.18 |
ENST00000545503.2 ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1 |
myosin binding protein C, slow type |
| chr2_-_96700664 | 0.18 |
ENST00000359548.4 ENST00000377137.3 ENST00000439254.1 ENST00000453542.1 |
GPAT2 |
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr22_-_32767017 | 0.18 |
ENST00000400234.1 |
RFPL3S |
RFPL3 antisense |
| chr11_-_117748138 | 0.18 |
ENST00000527717.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr5_-_95297678 | 0.18 |
ENST00000237853.4 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr22_-_32766972 | 0.18 |
ENST00000382084.4 ENST00000382086.2 |
RFPL3S |
RFPL3 antisense |
| chr16_-_29934558 | 0.18 |
ENST00000568995.1 ENST00000566413.1 |
KCTD13 |
potassium channel tetramerization domain containing 13 |
| chr16_+_12059050 | 0.18 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr9_-_95166884 | 0.17 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr6_+_52051171 | 0.17 |
ENST00000340057.1 |
IL17A |
interleukin 17A |
| chr6_+_26402517 | 0.17 |
ENST00000414912.2 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chr14_+_104182105 | 0.17 |
ENST00000311141.2 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr19_-_14606900 | 0.17 |
ENST00000393029.3 ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
| chr4_-_72649763 | 0.16 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr7_-_121944491 | 0.16 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr6_-_111927062 | 0.16 |
ENST00000359831.4 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr3_+_190231891 | 0.16 |
ENST00000434491.1 ENST00000422940.1 ENST00000317757.3 |
IL1RAP |
interleukin 1 receptor accessory protein |
| chr20_-_50418972 | 0.16 |
ENST00000395997.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr20_-_50418947 | 0.16 |
ENST00000371539.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr6_-_27100529 | 0.16 |
ENST00000607124.1 ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ |
histone cluster 1, H2bj |
| chr3_-_47324242 | 0.16 |
ENST00000456548.1 ENST00000432493.1 ENST00000335044.2 ENST00000444589.2 |
KIF9 |
kinesin family member 9 |
| chr2_+_10091815 | 0.16 |
ENST00000324907.9 |
GRHL1 |
grainyhead-like 1 (Drosophila) |
| chr19_+_45394477 | 0.16 |
ENST00000252487.5 ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr2_+_90211643 | 0.16 |
ENST00000390277.2 |
IGKV3D-11 |
immunoglobulin kappa variable 3D-11 |
| chr3_-_112693865 | 0.15 |
ENST00000471858.1 ENST00000295863.4 ENST00000308611.3 |
CD200R1 |
CD200 receptor 1 |
| chr21_-_34185989 | 0.15 |
ENST00000487113.1 ENST00000382373.4 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr20_-_50419055 | 0.15 |
ENST00000217086.4 |
SALL4 |
spalt-like transcription factor 4 |
| chr7_+_115862858 | 0.15 |
ENST00000393481.2 |
TES |
testis derived transcript (3 LIM domains) |
| chr2_+_182850743 | 0.15 |
ENST00000409702.1 |
PPP1R1C |
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr10_-_118928543 | 0.15 |
ENST00000419373.2 |
RP11-501J20.2 |
RP11-501J20.2 |
| chr1_-_185597619 | 0.15 |
ENST00000608417.1 ENST00000436955.1 |
GS1-204I12.1 |
GS1-204I12.1 |
| chr6_-_170151603 | 0.15 |
ENST00000366774.3 |
TCTE3 |
t-complex-associated-testis-expressed 3 |
| chr12_-_22063787 | 0.15 |
ENST00000544039.1 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr1_+_196857144 | 0.15 |
ENST00000367416.2 ENST00000251424.4 ENST00000367418.2 |
CFHR4 |
complement factor H-related 4 |
| chr2_-_69180083 | 0.15 |
ENST00000328895.4 |
GKN2 |
gastrokine 2 |
| chr3_+_121774202 | 0.14 |
ENST00000469710.1 ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86 |
CD86 molecule |
| chr1_+_12976450 | 0.14 |
ENST00000361079.2 |
PRAMEF7 |
PRAME family member 7 |
| chr4_+_155484155 | 0.14 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr10_+_5135981 | 0.14 |
ENST00000380554.3 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
| chr22_-_29107919 | 0.14 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr2_-_89247338 | 0.14 |
ENST00000496168.1 |
IGKV1-5 |
immunoglobulin kappa variable 1-5 |
| chr2_+_90077680 | 0.14 |
ENST00000390270.2 |
IGKV3D-20 |
immunoglobulin kappa variable 3D-20 |
| chr7_+_144052381 | 0.13 |
ENST00000498580.1 ENST00000056217.5 |
ARHGEF5 |
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr15_-_55489097 | 0.13 |
ENST00000260443.4 |
RSL24D1 |
ribosomal L24 domain containing 1 |
| chr4_-_138453559 | 0.13 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr10_-_112064665 | 0.13 |
ENST00000369603.5 |
SMNDC1 |
survival motor neuron domain containing 1 |
| chr7_-_87856280 | 0.13 |
ENST00000490437.1 ENST00000431660.1 |
SRI |
sorcin |
| chr4_+_74347400 | 0.13 |
ENST00000226355.3 |
AFM |
afamin |
| chr1_+_152974218 | 0.13 |
ENST00000331860.3 ENST00000443178.1 ENST00000295367.4 |
SPRR3 |
small proline-rich protein 3 |
| chr7_-_87856303 | 0.13 |
ENST00000394641.3 |
SRI |
sorcin |
| chr6_+_3259148 | 0.13 |
ENST00000419065.2 ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4 |
proteasome (prosome, macropain) assembly chaperone 4 |
| chr1_+_151739131 | 0.13 |
ENST00000400999.1 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr11_-_22647350 | 0.13 |
ENST00000327470.3 |
FANCF |
Fanconi anemia, complementation group F |
| chr1_-_17766198 | 0.13 |
ENST00000375436.4 |
RCC2 |
regulator of chromosome condensation 2 |
| chr18_-_3219847 | 0.12 |
ENST00000261606.7 |
MYOM1 |
myomesin 1 |
| chr6_-_26124138 | 0.12 |
ENST00000314332.5 ENST00000396984.1 |
HIST1H2BC |
histone cluster 1, H2bc |
| chr7_-_25268104 | 0.12 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chrX_+_105192423 | 0.12 |
ENST00000540278.1 |
NRK |
Nik related kinase |
| chr6_-_711395 | 0.12 |
ENST00000606285.1 |
RP11-532F6.3 |
RP11-532F6.3 |
| chr1_+_220267429 | 0.12 |
ENST00000366922.1 ENST00000302637.5 |
IARS2 |
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr22_+_17956618 | 0.12 |
ENST00000262608.8 |
CECR2 |
cat eye syndrome chromosome region, candidate 2 |
| chrX_+_107288239 | 0.12 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chrX_+_37639302 | 0.12 |
ENST00000545017.1 ENST00000536160.1 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr14_-_74959978 | 0.12 |
ENST00000541064.1 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr2_+_26568965 | 0.12 |
ENST00000260585.7 ENST00000447170.1 |
EPT1 |
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr14_+_56584414 | 0.12 |
ENST00000559044.1 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
| chr14_-_20801427 | 0.12 |
ENST00000557665.1 ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1 |
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr1_+_1260147 | 0.12 |
ENST00000343938.4 |
GLTPD1 |
glycolipid transfer protein domain containing 1 |
| chr14_-_74960030 | 0.12 |
ENST00000553490.1 ENST00000557510.1 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr4_+_5527117 | 0.12 |
ENST00000505296.1 |
C4orf6 |
chromosome 4 open reading frame 6 |
| chr7_+_93535866 | 0.12 |
ENST00000429473.1 ENST00000430875.1 ENST00000428834.1 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr3_-_129147432 | 0.11 |
ENST00000503957.1 ENST00000505956.1 ENST00000326085.3 |
EFCAB12 |
EF-hand calcium binding domain 12 |
| chr9_-_88897426 | 0.11 |
ENST00000375991.4 ENST00000326094.4 |
ISCA1 |
iron-sulfur cluster assembly 1 |
| chr2_-_40006289 | 0.11 |
ENST00000260619.6 ENST00000454352.2 |
THUMPD2 |
THUMP domain containing 2 |
| chr4_-_120243545 | 0.11 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr4_+_5526883 | 0.11 |
ENST00000195455.2 |
C4orf6 |
chromosome 4 open reading frame 6 |
| chr6_-_111927449 | 0.11 |
ENST00000368761.5 ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr1_-_201140673 | 0.11 |
ENST00000367333.2 |
TMEM9 |
transmembrane protein 9 |
| chrX_-_48271344 | 0.11 |
ENST00000376884.2 ENST00000396928.1 |
SSX4B |
synovial sarcoma, X breakpoint 4B |
| chr11_+_122753391 | 0.11 |
ENST00000307257.6 ENST00000227349.2 |
C11orf63 |
chromosome 11 open reading frame 63 |
| chr11_-_128894053 | 0.11 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr2_-_89327228 | 0.11 |
ENST00000483158.1 |
IGKV3-11 |
immunoglobulin kappa variable 3-11 |
| chr19_-_3557570 | 0.11 |
ENST00000355415.2 |
MFSD12 |
major facilitator superfamily domain containing 12 |
| chr5_+_40841276 | 0.11 |
ENST00000254691.5 |
CARD6 |
caspase recruitment domain family, member 6 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.3 | 1.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 2.5 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.2 | 0.7 | GO:0030336 | negative regulation of cell migration(GO:0030336) negative regulation of locomotion(GO:0040013) negative regulation of cellular component movement(GO:0051271) negative regulation of cell motility(GO:2000146) |
| 0.1 | 0.4 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.8 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.5 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.5 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.3 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.1 | 0.5 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.1 | 1.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.3 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.4 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.4 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.5 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.7 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.2 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.2 | GO:0043132 | coenzyme A transport(GO:0015880) FAD transport(GO:0015883) coenzyme A transmembrane transport(GO:0035349) FAD transmembrane transport(GO:0035350) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.1 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:2001186 | negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) |
| 0.0 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.4 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 1.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 1.0 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:1903301 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.4 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.4 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.3 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.3 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.0 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.3 | GO:0072025 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:1904247 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0072108 | positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) |
| 0.0 | 0.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.7 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 1.0 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.6 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.2 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0051971 | negative regulation of transmission of nerve impulse(GO:0051970) positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 | 0.1 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.3 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.0 | 0.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.0 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.8 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 0.4 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.6 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 1.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.9 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.0 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.4 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.1 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.4 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.2 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) FAD transmembrane transporter activity(GO:0015230) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.3 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 1.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.4 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.1 | 0.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 1.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 2.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.5 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 4.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |