Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for PGR
Z-value: 0.94
Transcription factors associated with PGR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PGR
|
ENSG00000082175.10 | PGR |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PGR | hg19_v2_chr11_-_100999775_100999801, hg19_v2_chr11_-_101000445_101000465 | 0.32 | 4.4e-01 | Click! |
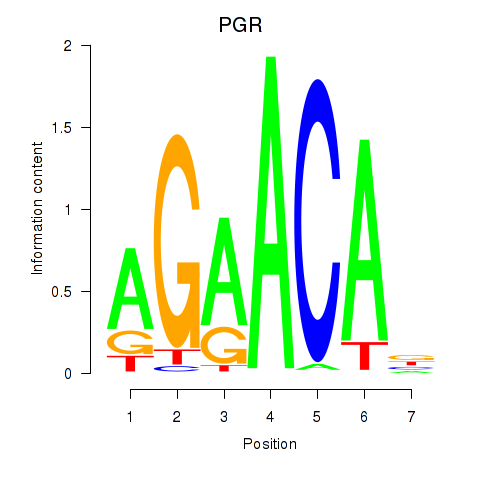
Activity profile of PGR motif
Sorted Z-values of PGR motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PGR
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_102104980 | 3.17 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr13_+_102104952 | 2.85 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr3_-_114343768 | 1.20 |
ENST00000393785.2 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr2_-_190044480 | 1.16 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr10_+_69865866 | 1.13 |
ENST00000354393.2 |
MYPN |
myopalladin |
| chrX_+_16964985 | 1.09 |
ENST00000303843.7 |
REPS2 |
RALBP1 associated Eps domain containing 2 |
| chr4_+_14113592 | 0.97 |
ENST00000502759.1 ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085 |
long intergenic non-protein coding RNA 1085 |
| chr14_-_61124977 | 0.86 |
ENST00000554986.1 |
SIX1 |
SIX homeobox 1 |
| chr3_-_114035026 | 0.77 |
ENST00000570269.1 |
RP11-553L6.5 |
RP11-553L6.5 |
| chr16_-_3350614 | 0.74 |
ENST00000268674.2 |
TIGD7 |
tigger transposable element derived 7 |
| chr3_-_114343039 | 0.67 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr6_+_72926145 | 0.67 |
ENST00000425662.2 ENST00000453976.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr1_-_85870177 | 0.66 |
ENST00000542148.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr13_+_32838801 | 0.65 |
ENST00000542859.1 |
FRY |
furry homolog (Drosophila) |
| chr1_+_155829286 | 0.65 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr3_-_58563094 | 0.58 |
ENST00000464064.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr1_-_145039835 | 0.58 |
ENST00000533259.1 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr5_+_67584174 | 0.58 |
ENST00000320694.8 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr2_-_175711133 | 0.57 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr7_-_150946015 | 0.56 |
ENST00000262188.8 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr10_-_49482907 | 0.54 |
ENST00000374201.3 ENST00000407470.4 |
FRMPD2 |
FERM and PDZ domain containing 2 |
| chr12_-_91505608 | 0.52 |
ENST00000266718.4 |
LUM |
lumican |
| chr6_-_43276535 | 0.51 |
ENST00000372569.3 ENST00000274990.4 |
CRIP3 |
cysteine-rich protein 3 |
| chrX_+_54834159 | 0.51 |
ENST00000375053.2 ENST00000347546.4 ENST00000375062.4 |
MAGED2 |
melanoma antigen family D, 2 |
| chr10_-_28571015 | 0.51 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr12_-_88974236 | 0.50 |
ENST00000228280.5 ENST00000552044.1 ENST00000357116.4 |
KITLG |
KIT ligand |
| chr6_-_154831779 | 0.49 |
ENST00000607772.1 |
CNKSR3 |
CNKSR family member 3 |
| chr5_-_121413974 | 0.49 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chr16_+_86600857 | 0.48 |
ENST00000320354.4 |
FOXC2 |
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
| chr1_+_78470530 | 0.46 |
ENST00000370763.5 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chrX_+_54834004 | 0.45 |
ENST00000375068.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr12_+_79258547 | 0.44 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chrX_+_57618269 | 0.44 |
ENST00000374888.1 |
ZXDB |
zinc finger, X-linked, duplicated B |
| chr11_+_20044600 | 0.43 |
ENST00000311043.8 |
NAV2 |
neuron navigator 2 |
| chr1_-_145039949 | 0.42 |
ENST00000313382.9 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr19_+_30863271 | 0.42 |
ENST00000355537.3 |
ZNF536 |
zinc finger protein 536 |
| chr20_+_23420322 | 0.41 |
ENST00000347397.1 |
CSTL1 |
cystatin-like 1 |
| chr5_-_124081008 | 0.40 |
ENST00000306315.5 |
ZNF608 |
zinc finger protein 608 |
| chr20_+_23420885 | 0.39 |
ENST00000246020.2 |
CSTL1 |
cystatin-like 1 |
| chr3_+_29323043 | 0.38 |
ENST00000452462.1 ENST00000456853.1 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
| chr10_+_104178946 | 0.38 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr8_-_7220490 | 0.37 |
ENST00000400078.2 |
ZNF705G |
zinc finger protein 705G |
| chr14_-_35182994 | 0.36 |
ENST00000341223.3 |
CFL2 |
cofilin 2 (muscle) |
| chr1_+_192778161 | 0.36 |
ENST00000235382.5 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
| chr5_+_137419581 | 0.36 |
ENST00000506684.1 ENST00000504809.1 ENST00000398754.1 |
WNT8A |
wingless-type MMTV integration site family, member 8A |
| chr1_-_156828810 | 0.36 |
ENST00000368195.3 |
INSRR |
insulin receptor-related receptor |
| chr2_+_10560147 | 0.35 |
ENST00000422133.1 |
HPCAL1 |
hippocalcin-like 1 |
| chr5_-_160279207 | 0.35 |
ENST00000327245.5 |
ATP10B |
ATPase, class V, type 10B |
| chr17_-_56621665 | 0.34 |
ENST00000321691.3 |
C17orf47 |
chromosome 17 open reading frame 47 |
| chr5_+_139505520 | 0.34 |
ENST00000333305.3 |
IGIP |
IgA-inducing protein |
| chr3_-_49722523 | 0.34 |
ENST00000448220.1 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr15_+_31658349 | 0.33 |
ENST00000558844.1 |
KLF13 |
Kruppel-like factor 13 |
| chr5_+_133861790 | 0.33 |
ENST00000395003.1 |
PHF15 |
jade family PHD finger 2 |
| chr16_-_66952779 | 0.33 |
ENST00000570262.1 ENST00000394055.3 ENST00000299752.4 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr1_-_179834311 | 0.33 |
ENST00000553856.1 |
IFRG15 |
Homo sapiens torsin A interacting protein 2 (TOR1AIP2), transcript variant 1, mRNA. |
| chr16_-_66952742 | 0.32 |
ENST00000565235.2 ENST00000568632.1 ENST00000565796.1 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr7_+_130126012 | 0.31 |
ENST00000341441.5 |
MEST |
mesoderm specific transcript |
| chr9_+_82188077 | 0.30 |
ENST00000425506.1 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr7_-_14026063 | 0.29 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr19_-_10446449 | 0.29 |
ENST00000592439.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr14_-_107219365 | 0.29 |
ENST00000424969.2 |
IGHV3-74 |
immunoglobulin heavy variable 3-74 |
| chr12_-_71551652 | 0.28 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr18_+_6729698 | 0.27 |
ENST00000383472.4 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr22_+_38219389 | 0.27 |
ENST00000249041.2 |
GALR3 |
galanin receptor 3 |
| chr4_+_15004165 | 0.27 |
ENST00000538197.1 ENST00000541112.1 ENST00000442003.2 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
| chr15_+_93447675 | 0.27 |
ENST00000536619.1 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr2_-_220034745 | 0.26 |
ENST00000455516.2 ENST00000396775.3 ENST00000295738.7 |
SLC23A3 |
solute carrier family 23, member 3 |
| chr19_-_48059113 | 0.26 |
ENST00000391901.3 ENST00000314121.4 ENST00000448976.1 |
ZNF541 |
zinc finger protein 541 |
| chr7_+_73442422 | 0.26 |
ENST00000358929.4 ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN |
elastin |
| chr5_+_133842243 | 0.26 |
ENST00000515627.2 |
AC005355.2 |
AC005355.2 |
| chr17_-_15168624 | 0.26 |
ENST00000312280.3 ENST00000494511.1 ENST00000580584.1 |
PMP22 |
peripheral myelin protein 22 |
| chr7_-_14026123 | 0.26 |
ENST00000420159.2 ENST00000399357.3 ENST00000403527.1 |
ETV1 |
ets variant 1 |
| chr9_-_113761720 | 0.25 |
ENST00000541779.1 ENST00000374430.2 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr7_+_73442487 | 0.25 |
ENST00000380575.4 ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN |
elastin |
| chr6_-_24489842 | 0.25 |
ENST00000230036.1 |
GPLD1 |
glycosylphosphatidylinositol specific phospholipase D1 |
| chr9_+_127615733 | 0.25 |
ENST00000373574.1 |
WDR38 |
WD repeat domain 38 |
| chr1_-_32210275 | 0.25 |
ENST00000440175.2 |
BAI2 |
brain-specific angiogenesis inhibitor 2 |
| chr11_-_73693875 | 0.25 |
ENST00000536983.1 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr2_-_85641162 | 0.25 |
ENST00000447219.2 ENST00000409670.1 ENST00000409724.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr19_+_41119323 | 0.25 |
ENST00000599724.1 ENST00000597071.1 ENST00000243562.9 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr3_-_151102529 | 0.25 |
ENST00000302632.3 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
| chr3_+_42695176 | 0.25 |
ENST00000232974.6 ENST00000457842.3 |
ZBTB47 |
zinc finger and BTB domain containing 47 |
| chr21_+_17566643 | 0.24 |
ENST00000419952.1 ENST00000445461.2 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr10_+_124030819 | 0.24 |
ENST00000260723.4 ENST00000368994.2 |
BTBD16 |
BTB (POZ) domain containing 16 |
| chr1_+_154975258 | 0.23 |
ENST00000417934.2 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
| chr7_+_73442457 | 0.23 |
ENST00000438880.1 ENST00000414324.1 ENST00000380562.4 |
ELN |
elastin |
| chr2_-_183387430 | 0.23 |
ENST00000410103.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr6_+_39760129 | 0.23 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr22_-_31688381 | 0.23 |
ENST00000487265.2 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chr7_-_23510086 | 0.22 |
ENST00000258729.3 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
| chr9_-_73736511 | 0.22 |
ENST00000377110.3 ENST00000377111.2 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr2_-_27531313 | 0.22 |
ENST00000296099.2 |
UCN |
urocortin |
| chr7_+_130126165 | 0.21 |
ENST00000427521.1 ENST00000416162.2 ENST00000378576.4 |
MEST |
mesoderm specific transcript |
| chr12_+_56661461 | 0.21 |
ENST00000546544.1 ENST00000553234.1 |
COQ10A |
coenzyme Q10 homolog A (S. cerevisiae) |
| chr9_-_131940526 | 0.21 |
ENST00000372491.2 |
IER5L |
immediate early response 5-like |
| chr9_-_34665983 | 0.20 |
ENST00000416454.1 ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5 |
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr11_+_6502675 | 0.19 |
ENST00000254616.6 ENST00000530751.1 |
TIMM10B |
translocase of inner mitochondrial membrane 10 homolog B (yeast) |
| chr2_-_216300784 | 0.19 |
ENST00000421182.1 ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1 |
fibronectin 1 |
| chr22_-_30162924 | 0.19 |
ENST00000344318.3 ENST00000397781.3 |
ZMAT5 |
zinc finger, matrin-type 5 |
| chr9_+_131683174 | 0.19 |
ENST00000372592.3 ENST00000428610.1 |
PHYHD1 |
phytanoyl-CoA dioxygenase domain containing 1 |
| chrX_+_135570046 | 0.19 |
ENST00000370648.3 |
BRS3 |
bombesin-like receptor 3 |
| chr2_+_42104692 | 0.19 |
ENST00000398796.2 ENST00000442214.1 |
AC104654.1 |
AC104654.1 |
| chr3_+_137906109 | 0.19 |
ENST00000481646.1 ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr17_+_57642886 | 0.19 |
ENST00000251241.4 ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr7_+_30960915 | 0.19 |
ENST00000441328.2 ENST00000409899.1 ENST00000409611.1 |
AQP1 |
aquaporin 1 (Colton blood group) |
| chr5_+_102201430 | 0.19 |
ENST00000438793.3 ENST00000346918.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr2_-_179315786 | 0.19 |
ENST00000457633.1 ENST00000438687.3 ENST00000325748.4 |
PRKRA |
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr13_+_24144509 | 0.18 |
ENST00000248484.4 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr15_+_94899183 | 0.18 |
ENST00000557742.1 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
| chr9_-_15510989 | 0.18 |
ENST00000380715.1 ENST00000380716.4 ENST00000380738.4 ENST00000380733.4 |
PSIP1 |
PC4 and SFRS1 interacting protein 1 |
| chr1_-_110933663 | 0.18 |
ENST00000369781.4 ENST00000541986.1 ENST00000369779.4 |
SLC16A4 |
solute carrier family 16, member 4 |
| chr1_-_167906277 | 0.18 |
ENST00000271373.4 |
MPC2 |
mitochondrial pyruvate carrier 2 |
| chr6_+_63921399 | 0.18 |
ENST00000356170.3 |
FKBP1C |
FK506 binding protein 1C |
| chr2_-_220034712 | 0.18 |
ENST00000409370.2 ENST00000430764.1 ENST00000409878.3 |
SLC23A3 |
solute carrier family 23, member 3 |
| chr2_-_158300556 | 0.18 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr6_+_108487245 | 0.18 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chr17_-_40540586 | 0.18 |
ENST00000264657.5 |
STAT3 |
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr19_+_13135790 | 0.18 |
ENST00000358552.3 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr6_-_150212029 | 0.17 |
ENST00000529948.1 ENST00000357183.4 ENST00000367363.3 |
RAET1E |
retinoic acid early transcript 1E |
| chr5_-_138718973 | 0.17 |
ENST00000353963.3 ENST00000348729.3 |
SLC23A1 |
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr13_+_49822041 | 0.17 |
ENST00000538056.1 ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1 |
cytidine and dCMP deaminase domain containing 1 |
| chr17_+_45286387 | 0.16 |
ENST00000572316.1 ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr3_-_52001448 | 0.16 |
ENST00000461554.1 ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4 |
poly(rC) binding protein 4 |
| chr17_+_45286706 | 0.16 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr2_-_188312971 | 0.16 |
ENST00000410068.1 ENST00000447403.1 ENST00000410102.1 |
CALCRL |
calcitonin receptor-like |
| chr2_-_179315453 | 0.16 |
ENST00000432031.2 |
PRKRA |
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr10_-_48416849 | 0.16 |
ENST00000249598.1 |
GDF2 |
growth differentiation factor 2 |
| chr7_+_22766766 | 0.16 |
ENST00000426291.1 ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6 |
interleukin 6 (interferon, beta 2) |
| chr3_+_52454971 | 0.16 |
ENST00000465863.1 |
PHF7 |
PHD finger protein 7 |
| chr11_-_124806297 | 0.16 |
ENST00000298251.4 |
HEPACAM |
hepatic and glial cell adhesion molecule |
| chr3_+_158991025 | 0.16 |
ENST00000337808.6 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr18_+_43304092 | 0.16 |
ENST00000321925.4 ENST00000587601.1 |
SLC14A1 |
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr5_+_102201509 | 0.15 |
ENST00000348126.2 ENST00000379787.4 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chrX_-_69509738 | 0.15 |
ENST00000374454.1 ENST00000239666.4 |
PDZD11 |
PDZ domain containing 11 |
| chr14_-_106692191 | 0.15 |
ENST00000390607.2 |
IGHV3-21 |
immunoglobulin heavy variable 3-21 |
| chr2_-_219925189 | 0.15 |
ENST00000295731.6 |
IHH |
indian hedgehog |
| chr1_-_154943002 | 0.15 |
ENST00000606391.1 |
SHC1 |
SHC (Src homology 2 domain containing) transforming protein 1 |
| chrX_+_106163626 | 0.15 |
ENST00000336803.1 |
CLDN2 |
claudin 2 |
| chr1_-_154943212 | 0.15 |
ENST00000368445.5 ENST00000448116.2 ENST00000368449.4 |
SHC1 |
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr19_+_507299 | 0.15 |
ENST00000359315.5 |
TPGS1 |
tubulin polyglutamylase complex subunit 1 |
| chr19_-_36247910 | 0.15 |
ENST00000587965.1 ENST00000004982.3 |
HSPB6 |
heat shock protein, alpha-crystallin-related, B6 |
| chr10_-_104178857 | 0.15 |
ENST00000020673.5 |
PSD |
pleckstrin and Sec7 domain containing |
| chr5_-_131347501 | 0.14 |
ENST00000543479.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr15_+_54305101 | 0.14 |
ENST00000260323.11 ENST00000545554.1 ENST00000537900.1 |
UNC13C |
unc-13 homolog C (C. elegans) |
| chr17_-_2966901 | 0.14 |
ENST00000575751.1 |
OR1D5 |
olfactory receptor, family 1, subfamily D, member 5 |
| chr21_+_17791648 | 0.14 |
ENST00000602892.1 ENST00000418813.2 ENST00000435697.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr8_+_9953214 | 0.14 |
ENST00000382490.5 |
MSRA |
methionine sulfoxide reductase A |
| chr2_-_179315490 | 0.14 |
ENST00000487082.1 |
PRKRA |
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr4_+_146539415 | 0.14 |
ENST00000281317.5 |
MMAA |
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr22_-_31688431 | 0.14 |
ENST00000402249.3 ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chr1_+_145516560 | 0.14 |
ENST00000537888.1 |
PEX11B |
peroxisomal biogenesis factor 11 beta |
| chrX_+_100743031 | 0.14 |
ENST00000423738.3 |
ARMCX4 |
armadillo repeat containing, X-linked 4 |
| chrX_-_114953669 | 0.14 |
ENST00000449327.1 |
RP1-241P17.4 |
Uncharacterized protein |
| chr17_+_75447326 | 0.14 |
ENST00000591088.1 |
SEPT9 |
septin 9 |
| chr6_-_43021437 | 0.14 |
ENST00000265348.3 |
CUL7 |
cullin 7 |
| chr1_-_145039771 | 0.13 |
ENST00000493130.2 ENST00000532801.1 ENST00000478649.2 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr5_+_140797296 | 0.13 |
ENST00000398594.2 |
PCDHGB7 |
protocadherin gamma subfamily B, 7 |
| chr2_-_163695128 | 0.13 |
ENST00000332142.5 |
KCNH7 |
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr18_+_13611763 | 0.13 |
ENST00000585931.1 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chr20_+_46130619 | 0.13 |
ENST00000372004.3 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr8_-_42623747 | 0.13 |
ENST00000534622.1 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr9_-_215744 | 0.13 |
ENST00000382387.2 |
C9orf66 |
chromosome 9 open reading frame 66 |
| chr17_+_73997796 | 0.13 |
ENST00000586261.1 |
CDK3 |
cyclin-dependent kinase 3 |
| chr1_+_155146318 | 0.13 |
ENST00000368385.4 ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46 |
tripartite motif containing 46 |
| chr12_-_53901266 | 0.13 |
ENST00000609999.1 ENST00000267017.3 |
NPFF |
neuropeptide FF-amide peptide precursor |
| chr3_+_185046676 | 0.13 |
ENST00000428617.1 ENST00000443863.1 |
MAP3K13 |
mitogen-activated protein kinase kinase kinase 13 |
| chr9_+_2159850 | 0.13 |
ENST00000416751.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_-_154164534 | 0.13 |
ENST00000271850.7 ENST00000368530.2 |
TPM3 |
tropomyosin 3 |
| chr18_+_6729725 | 0.12 |
ENST00000400091.2 ENST00000583410.1 ENST00000584387.1 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr17_-_42988004 | 0.12 |
ENST00000586125.1 ENST00000591880.1 |
GFAP |
glial fibrillary acidic protein |
| chr7_+_73442102 | 0.12 |
ENST00000445912.1 ENST00000252034.7 |
ELN |
elastin |
| chr6_-_88299678 | 0.12 |
ENST00000369536.5 |
RARS2 |
arginyl-tRNA synthetase 2, mitochondrial |
| chr3_+_44626446 | 0.12 |
ENST00000441021.1 ENST00000322734.2 |
ZNF660 |
zinc finger protein 660 |
| chr4_-_152149033 | 0.11 |
ENST00000514152.1 |
SH3D19 |
SH3 domain containing 19 |
| chr11_-_615570 | 0.11 |
ENST00000525445.1 ENST00000348655.6 ENST00000397566.1 |
IRF7 |
interferon regulatory factor 7 |
| chr11_-_6502534 | 0.11 |
ENST00000254584.2 ENST00000525235.1 ENST00000445086.2 |
ARFIP2 |
ADP-ribosylation factor interacting protein 2 |
| chr17_+_37824411 | 0.11 |
ENST00000269582.2 |
PNMT |
phenylethanolamine N-methyltransferase |
| chr17_+_46970134 | 0.11 |
ENST00000503641.1 ENST00000514808.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr19_+_13135386 | 0.11 |
ENST00000360105.4 ENST00000588228.1 ENST00000591028.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr11_-_75379612 | 0.11 |
ENST00000526740.1 |
MAP6 |
microtubule-associated protein 6 |
| chr5_-_131347583 | 0.11 |
ENST00000379255.1 ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr13_-_103053946 | 0.11 |
ENST00000376131.4 |
FGF14 |
fibroblast growth factor 14 |
| chr4_-_154710210 | 0.11 |
ENST00000274063.4 |
SFRP2 |
secreted frizzled-related protein 2 |
| chr7_+_134576151 | 0.11 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr13_+_43148281 | 0.11 |
ENST00000239849.6 ENST00000398795.2 ENST00000544862.1 |
TNFSF11 |
tumor necrosis factor (ligand) superfamily, member 11 |
| chr20_-_44168046 | 0.11 |
ENST00000372665.3 ENST00000372670.3 ENST00000600168.1 |
WFDC6 |
WAP four-disulfide core domain 6 |
| chr11_-_6502580 | 0.11 |
ENST00000423813.2 ENST00000396777.3 |
ARFIP2 |
ADP-ribosylation factor interacting protein 2 |
| chr15_+_67841330 | 0.11 |
ENST00000354498.5 |
MAP2K5 |
mitogen-activated protein kinase kinase 5 |
| chr1_-_115292591 | 0.11 |
ENST00000438362.2 |
CSDE1 |
cold shock domain containing E1, RNA-binding |
| chr1_+_153232160 | 0.11 |
ENST00000368742.3 |
LOR |
loricrin |
| chr2_+_9778872 | 0.10 |
ENST00000478468.1 |
RP11-521D12.1 |
RP11-521D12.1 |
| chr12_+_53848505 | 0.10 |
ENST00000552819.1 ENST00000455667.3 |
PCBP2 |
poly(rC) binding protein 2 |
| chrX_-_21776281 | 0.10 |
ENST00000379494.3 |
SMPX |
small muscle protein, X-linked |
| chr9_+_71394945 | 0.10 |
ENST00000394264.3 |
FAM122A |
family with sequence similarity 122A |
| chr17_-_76183111 | 0.10 |
ENST00000405273.1 ENST00000590862.1 ENST00000590430.1 ENST00000586613.1 |
TK1 |
thymidine kinase 1, soluble |
| chr19_+_44488330 | 0.10 |
ENST00000591532.1 ENST00000407951.2 ENST00000270014.2 ENST00000590615.1 ENST00000586454.1 |
ZNF155 |
zinc finger protein 155 |
| chr5_+_102201687 | 0.10 |
ENST00000304400.7 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr6_-_76782371 | 0.10 |
ENST00000369950.3 ENST00000369963.3 |
IMPG1 |
interphotoreceptor matrix proteoglycan 1 |
| chr8_+_10530155 | 0.10 |
ENST00000521818.1 |
C8orf74 |
chromosome 8 open reading frame 74 |
| chr17_+_46970178 | 0.10 |
ENST00000393366.2 ENST00000506855.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.7 | GO:1905154 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 0.9 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 0.5 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.2 | 0.5 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) positive regulation of vascular wound healing(GO:0035470) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.2 | 0.2 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.4 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.5 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.6 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 1.0 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.7 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 0.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.4 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.1 | 0.3 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.3 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.2 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 | 0.3 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.2 | GO:0072229 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 1.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.2 | GO:0070837 | L-ascorbic acid transport(GO:0015882) dehydroascorbic acid transport(GO:0070837) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.2 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.1 | GO:0042662 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.2 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.2 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.7 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.2 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.4 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.0 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) |
| 0.0 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0100009 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.0 | 2.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:1904124 | negative regulation of norepinephrine secretion(GO:0010700) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.6 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.5 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.5 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0018095 | sperm axoneme assembly(GO:0007288) protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.0 | GO:1904798 | regulation of core promoter binding(GO:1904796) positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.6 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.4 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.2 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.5 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.5 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.5 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 0.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.6 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.2 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.2 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.1 | 0.2 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 0.2 | GO:0033300 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) dehydroascorbic acid transporter activity(GO:0033300) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 1.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.0 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.9 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 2.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.5 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |