Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for PITX3
Z-value: 0.20
Transcription factors associated with PITX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX3
|
ENSG00000107859.5 | PITX3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX3 | hg19_v2_chr10_-_104001231_104001274 | -0.22 | 5.9e-01 | Click! |
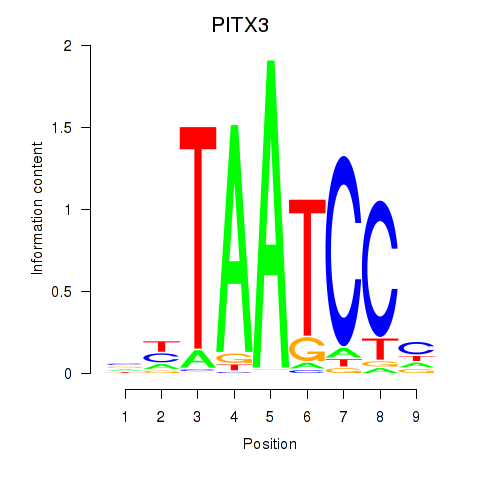
Activity profile of PITX3 motif
Sorted Z-values of PITX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_75766763 | 0.47 |
ENST00000456643.1 ENST00000415424.1 |
ANXA1 |
annexin A1 |
| chr1_+_163038565 | 0.43 |
ENST00000421743.2 |
RGS4 |
regulator of G-protein signaling 4 |
| chr3_-_149293990 | 0.34 |
ENST00000472417.1 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr1_+_12916941 | 0.33 |
ENST00000240189.2 |
PRAMEF2 |
PRAME family member 2 |
| chr14_-_106539557 | 0.33 |
ENST00000390599.2 |
IGHV1-8 |
immunoglobulin heavy variable 1-8 |
| chr3_-_112565703 | 0.31 |
ENST00000488794.1 |
CD200R1L |
CD200 receptor 1-like |
| chr7_+_36429424 | 0.29 |
ENST00000396068.2 |
ANLN |
anillin, actin binding protein |
| chr3_+_111393501 | 0.29 |
ENST00000393934.3 |
PLCXD2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr9_-_39288092 | 0.28 |
ENST00000323947.7 ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3 |
contactin associated protein-like 3 |
| chr21_+_25801041 | 0.28 |
ENST00000453784.2 ENST00000423581.1 |
AP000476.1 |
AP000476.1 |
| chr3_+_157154578 | 0.27 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr10_-_70231639 | 0.27 |
ENST00000551118.2 ENST00000358410.3 ENST00000399180.2 ENST00000399179.2 |
DNA2 |
DNA replication helicase/nuclease 2 |
| chr5_-_146781153 | 0.25 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr2_+_120770581 | 0.25 |
ENST00000263713.5 |
EPB41L5 |
erythrocyte membrane protein band 4.1 like 5 |
| chr7_+_44143925 | 0.24 |
ENST00000223357.3 |
AEBP1 |
AE binding protein 1 |
| chr13_+_102104980 | 0.23 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr12_+_51632666 | 0.22 |
ENST00000604900.1 |
DAZAP2 |
DAZ associated protein 2 |
| chr8_+_38261880 | 0.22 |
ENST00000527175.1 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr2_-_19558373 | 0.22 |
ENST00000272223.2 |
OSR1 |
odd-skipped related transciption factor 1 |
| chr1_+_144989309 | 0.22 |
ENST00000596396.1 |
AL590452.1 |
Uncharacterized protein |
| chr6_+_83073952 | 0.22 |
ENST00000543496.1 |
TPBG |
trophoblast glycoprotein |
| chr3_-_112564797 | 0.22 |
ENST00000398214.1 ENST00000448932.1 |
CD200R1L |
CD200 receptor 1-like |
| chrX_+_118425471 | 0.22 |
ENST00000428222.1 |
RP5-1139I1.1 |
RP5-1139I1.1 |
| chr6_+_10585979 | 0.21 |
ENST00000265012.4 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr4_+_126237554 | 0.21 |
ENST00000394329.3 |
FAT4 |
FAT atypical cadherin 4 |
| chr1_+_55505184 | 0.21 |
ENST00000302118.5 |
PCSK9 |
proprotein convertase subtilisin/kexin type 9 |
| chr7_+_36429409 | 0.21 |
ENST00000265748.2 |
ANLN |
anillin, actin binding protein |
| chr1_+_160765947 | 0.21 |
ENST00000263285.6 ENST00000368039.2 |
LY9 |
lymphocyte antigen 9 |
| chr12_+_60058458 | 0.21 |
ENST00000548610.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr9_-_95186739 | 0.21 |
ENST00000375550.4 |
OMD |
osteomodulin |
| chr2_-_202562774 | 0.21 |
ENST00000396886.3 ENST00000409143.1 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr14_-_106471723 | 0.20 |
ENST00000390595.2 |
IGHV1-3 |
immunoglobulin heavy variable 1-3 |
| chr2_-_202563414 | 0.20 |
ENST00000409474.3 ENST00000315506.7 ENST00000359962.5 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr4_+_154622652 | 0.20 |
ENST00000260010.6 |
TLR2 |
toll-like receptor 2 |
| chr1_+_12851545 | 0.20 |
ENST00000332296.7 |
PRAMEF1 |
PRAME family member 1 |
| chr19_+_54372639 | 0.20 |
ENST00000391769.2 |
MYADM |
myeloid-associated differentiation marker |
| chr12_+_52668394 | 0.20 |
ENST00000423955.2 |
KRT86 |
keratin 86 |
| chr8_-_124428569 | 0.19 |
ENST00000521903.1 |
ATAD2 |
ATPase family, AAA domain containing 2 |
| chr19_-_44160768 | 0.19 |
ENST00000593447.1 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr2_-_190044480 | 0.19 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chrM_+_5824 | 0.19 |
ENST00000361624.2 |
MT-CO1 |
mitochondrially encoded cytochrome c oxidase I |
| chr11_+_20044096 | 0.19 |
ENST00000533917.1 |
NAV2 |
neuron navigator 2 |
| chr5_-_41510725 | 0.19 |
ENST00000328457.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr13_-_25086879 | 0.19 |
ENST00000381989.3 |
PARP4 |
poly (ADP-ribose) polymerase family, member 4 |
| chr4_+_170541835 | 0.19 |
ENST00000504131.2 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr22_+_19467261 | 0.18 |
ENST00000455750.1 ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45 |
cell division cycle 45 |
| chrX_-_65259900 | 0.18 |
ENST00000412866.2 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr17_-_29648761 | 0.18 |
ENST00000247270.3 ENST00000462804.2 |
EVI2A |
ecotropic viral integration site 2A |
| chr19_-_15343773 | 0.18 |
ENST00000435261.1 ENST00000594042.1 |
EPHX3 |
epoxide hydrolase 3 |
| chr6_+_10556215 | 0.18 |
ENST00000316170.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr1_-_21606013 | 0.18 |
ENST00000357071.4 |
ECE1 |
endothelin converting enzyme 1 |
| chr19_-_44174330 | 0.18 |
ENST00000340093.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr11_-_62752162 | 0.18 |
ENST00000458333.2 ENST00000421062.2 |
SLC22A6 |
solute carrier family 22 (organic anion transporter), member 6 |
| chr3_+_73110810 | 0.18 |
ENST00000533473.1 |
EBLN2 |
endogenous Bornavirus-like nucleoprotein 2 |
| chr19_-_44174305 | 0.18 |
ENST00000601723.1 ENST00000339082.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr3_-_50340996 | 0.18 |
ENST00000266031.4 ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1 |
hyaluronoglucosaminidase 1 |
| chr18_-_70532906 | 0.17 |
ENST00000299430.2 ENST00000397929.1 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr14_+_58894103 | 0.17 |
ENST00000354386.6 ENST00000556134.1 |
KIAA0586 |
KIAA0586 |
| chr5_-_41510656 | 0.17 |
ENST00000377801.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chrX_+_100333709 | 0.17 |
ENST00000372930.4 |
TMEM35 |
transmembrane protein 35 |
| chrX_-_45629661 | 0.17 |
ENST00000602507.1 ENST00000602461.1 |
RP6-99M1.2 |
RP6-99M1.2 |
| chr12_+_32655048 | 0.17 |
ENST00000427716.2 ENST00000266482.3 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chrM_-_14670 | 0.17 |
ENST00000361681.2 |
MT-ND6 |
mitochondrially encoded NADH dehydrogenase 6 |
| chrX_-_65253506 | 0.17 |
ENST00000427538.1 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr1_+_93646238 | 0.17 |
ENST00000448243.1 ENST00000370276.1 |
CCDC18 |
coiled-coil domain containing 18 |
| chr10_-_129924468 | 0.17 |
ENST00000368653.3 |
MKI67 |
marker of proliferation Ki-67 |
| chr10_+_115674530 | 0.17 |
ENST00000451472.1 |
AL162407.1 |
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr19_+_39687596 | 0.17 |
ENST00000339852.4 |
NCCRP1 |
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr3_-_149095652 | 0.16 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr12_-_10978957 | 0.16 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr4_-_153601136 | 0.16 |
ENST00000504064.1 ENST00000304385.3 |
TMEM154 |
transmembrane protein 154 |
| chr6_+_31638156 | 0.16 |
ENST00000409525.1 |
LY6G5B |
lymphocyte antigen 6 complex, locus G5B |
| chr4_+_113970772 | 0.16 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr15_+_32933866 | 0.16 |
ENST00000300175.4 ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5 |
secretogranin V (7B2 protein) |
| chr10_-_49860525 | 0.16 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr8_-_131399110 | 0.16 |
ENST00000521426.1 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr2_-_89459813 | 0.16 |
ENST00000390256.2 |
IGKV6-21 |
immunoglobulin kappa variable 6-21 (non-functional) |
| chr1_+_207262881 | 0.16 |
ENST00000451804.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr8_+_40018977 | 0.16 |
ENST00000520487.1 |
RP11-470M17.2 |
RP11-470M17.2 |
| chr2_+_152214098 | 0.16 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr5_-_111312622 | 0.16 |
ENST00000395634.3 |
NREP |
neuronal regeneration related protein |
| chr2_-_40739501 | 0.15 |
ENST00000403092.1 ENST00000402441.1 ENST00000448531.1 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr19_+_39786962 | 0.15 |
ENST00000333625.2 |
IFNL1 |
interferon, lambda 1 |
| chr1_-_31230650 | 0.15 |
ENST00000294507.3 |
LAPTM5 |
lysosomal protein transmembrane 5 |
| chr14_+_101299520 | 0.15 |
ENST00000455531.1 |
MEG3 |
maternally expressed 3 (non-protein coding) |
| chr1_-_241799232 | 0.15 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chr14_+_58894141 | 0.15 |
ENST00000423743.3 |
KIAA0586 |
KIAA0586 |
| chrX_-_65259914 | 0.15 |
ENST00000374737.4 ENST00000455586.2 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr13_+_102104952 | 0.15 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr19_-_15344243 | 0.15 |
ENST00000602233.1 |
EPHX3 |
epoxide hydrolase 3 |
| chr5_-_16936340 | 0.15 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr4_-_171011323 | 0.15 |
ENST00000509167.1 ENST00000353187.2 ENST00000507375.1 ENST00000515480.1 |
AADAT |
aminoadipate aminotransferase |
| chr11_+_30252549 | 0.15 |
ENST00000254122.3 ENST00000417547.1 |
FSHB |
follicle stimulating hormone, beta polypeptide |
| chr9_-_35958151 | 0.15 |
ENST00000341959.2 |
OR2S2 |
olfactory receptor, family 2, subfamily S, member 2 |
| chr17_-_10633291 | 0.14 |
ENST00000578345.1 ENST00000455996.2 |
TMEM220 |
transmembrane protein 220 |
| chr7_+_48211048 | 0.14 |
ENST00000435803.1 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr8_+_27629459 | 0.14 |
ENST00000523566.1 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr1_+_45805342 | 0.14 |
ENST00000372090.5 |
TOE1 |
target of EGR1, member 1 (nuclear) |
| chr10_-_103578162 | 0.14 |
ENST00000361464.3 ENST00000357797.5 ENST00000370094.3 |
MGEA5 |
meningioma expressed antigen 5 (hyaluronidase) |
| chr8_+_66955648 | 0.14 |
ENST00000522619.1 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr15_-_56285742 | 0.14 |
ENST00000435532.3 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr11_+_61891445 | 0.14 |
ENST00000394818.3 ENST00000533896.1 ENST00000278849.4 |
INCENP |
inner centromere protein antigens 135/155kDa |
| chr13_-_31191642 | 0.14 |
ENST00000405805.1 |
HMGB1 |
high mobility group box 1 |
| chr1_-_150693318 | 0.14 |
ENST00000442853.1 ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1 |
HORMA domain containing 1 |
| chr6_+_10694900 | 0.14 |
ENST00000379568.3 |
PAK1IP1 |
PAK1 interacting protein 1 |
| chr9_+_71986182 | 0.14 |
ENST00000303068.7 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr5_-_38595498 | 0.14 |
ENST00000263409.4 |
LIFR |
leukemia inhibitory factor receptor alpha |
| chr15_-_98417780 | 0.14 |
ENST00000503874.3 |
LINC00923 |
long intergenic non-protein coding RNA 923 |
| chr3_+_10068095 | 0.14 |
ENST00000287647.3 ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2 |
Fanconi anemia, complementation group D2 |
| chr20_-_5485166 | 0.14 |
ENST00000589201.1 ENST00000379053.4 |
LINC00654 |
long intergenic non-protein coding RNA 654 |
| chr1_+_117602925 | 0.14 |
ENST00000369466.4 |
TTF2 |
transcription termination factor, RNA polymerase II |
| chr18_-_40857493 | 0.14 |
ENST00000255224.3 |
SYT4 |
synaptotagmin IV |
| chr19_-_2456922 | 0.14 |
ENST00000582871.1 ENST00000325327.3 |
LMNB2 |
lamin B2 |
| chr1_+_159409512 | 0.14 |
ENST00000423932.3 |
OR10J1 |
olfactory receptor, family 10, subfamily J, member 1 |
| chr8_+_25316707 | 0.14 |
ENST00000380665.3 |
CDCA2 |
cell division cycle associated 2 |
| chr8_+_41386725 | 0.14 |
ENST00000276533.3 ENST00000520710.1 ENST00000518671.1 |
GINS4 |
GINS complex subunit 4 (Sld5 homolog) |
| chr10_+_124320156 | 0.14 |
ENST00000338354.3 ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr12_-_4553385 | 0.14 |
ENST00000543077.1 |
FGF6 |
fibroblast growth factor 6 |
| chr10_-_106098162 | 0.14 |
ENST00000337478.1 |
ITPRIP |
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr2_-_85555355 | 0.14 |
ENST00000282120.2 ENST00000398263.2 |
TGOLN2 |
trans-golgi network protein 2 |
| chr1_+_114522049 | 0.14 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr14_+_22217447 | 0.13 |
ENST00000390427.3 |
TRAV5 |
T cell receptor alpha variable 5 |
| chr12_-_90049828 | 0.13 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr9_-_93405352 | 0.13 |
ENST00000375765.3 |
DIRAS2 |
DIRAS family, GTP-binding RAS-like 2 |
| chr3_+_141105705 | 0.13 |
ENST00000513258.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr17_-_8113542 | 0.13 |
ENST00000578549.1 ENST00000535053.1 ENST00000582368.1 |
AURKB |
aurora kinase B |
| chr9_+_134000948 | 0.13 |
ENST00000359428.5 ENST00000411637.2 ENST00000451030.1 |
NUP214 |
nucleoporin 214kDa |
| chr16_-_21877629 | 0.13 |
ENST00000544693.1 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chr1_+_214776516 | 0.13 |
ENST00000366955.3 |
CENPF |
centromere protein F, 350/400kDa |
| chr7_-_111846435 | 0.13 |
ENST00000437633.1 ENST00000428084.1 |
DOCK4 |
dedicator of cytokinesis 4 |
| chr10_+_70847852 | 0.13 |
ENST00000242465.3 |
SRGN |
serglycin |
| chr3_-_121264848 | 0.13 |
ENST00000264233.5 |
POLQ |
polymerase (DNA directed), theta |
| chr12_+_16500037 | 0.13 |
ENST00000536371.1 ENST00000010404.2 |
MGST1 |
microsomal glutathione S-transferase 1 |
| chrX_+_69509927 | 0.13 |
ENST00000374403.3 |
KIF4A |
kinesin family member 4A |
| chr4_-_120988229 | 0.13 |
ENST00000296509.6 |
MAD2L1 |
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr1_-_47407097 | 0.13 |
ENST00000457840.2 |
CYP4A11 |
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr17_-_10633535 | 0.13 |
ENST00000341871.3 |
TMEM220 |
transmembrane protein 220 |
| chr1_+_45205478 | 0.13 |
ENST00000452259.1 ENST00000372224.4 |
KIF2C |
kinesin family member 2C |
| chr14_-_107078851 | 0.13 |
ENST00000390628.2 |
IGHV1-58 |
immunoglobulin heavy variable 1-58 |
| chr3_-_20227619 | 0.13 |
ENST00000425061.1 ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1 |
shugoshin-like 1 (S. pombe) |
| chr22_+_31477296 | 0.13 |
ENST00000426927.1 ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN |
smoothelin |
| chr17_+_43318434 | 0.13 |
ENST00000587489.1 |
FMNL1 |
formin-like 1 |
| chr2_-_163100045 | 0.13 |
ENST00000188790.4 |
FAP |
fibroblast activation protein, alpha |
| chr7_-_138348969 | 0.12 |
ENST00000436657.1 |
SVOPL |
SVOP-like |
| chr10_-_129924611 | 0.12 |
ENST00000368654.3 |
MKI67 |
marker of proliferation Ki-67 |
| chr7_-_33080506 | 0.12 |
ENST00000381626.2 ENST00000409467.1 ENST00000449201.1 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr2_-_163099885 | 0.12 |
ENST00000443424.1 |
FAP |
fibroblast activation protein, alpha |
| chr1_+_246887349 | 0.12 |
ENST00000366510.3 |
SCCPDH |
saccharopine dehydrogenase (putative) |
| chr17_-_63822563 | 0.12 |
ENST00000317442.8 |
CEP112 |
centrosomal protein 112kDa |
| chr9_+_91605778 | 0.12 |
ENST00000375851.2 ENST00000375850.3 ENST00000334490.5 |
C9orf47 |
chromosome 9 open reading frame 47 |
| chr16_+_22516172 | 0.12 |
ENST00000543407.1 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr17_-_39274606 | 0.12 |
ENST00000391413.2 |
KRTAP4-11 |
keratin associated protein 4-11 |
| chr3_-_113918254 | 0.12 |
ENST00000460779.1 |
DRD3 |
dopamine receptor D3 |
| chr6_+_111580508 | 0.12 |
ENST00000368847.4 |
KIAA1919 |
KIAA1919 |
| chrM_+_9207 | 0.12 |
ENST00000362079.2 |
MT-CO3 |
mitochondrially encoded cytochrome c oxidase III |
| chr16_+_53133070 | 0.12 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr7_-_14026063 | 0.12 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr9_+_5510558 | 0.12 |
ENST00000397747.3 |
PDCD1LG2 |
programmed cell death 1 ligand 2 |
| chr19_-_19314162 | 0.12 |
ENST00000420605.3 ENST00000544883.1 ENST00000538165.2 ENST00000331552.7 |
NR2C2AP |
nuclear receptor 2C2-associated protein |
| chr4_-_107957454 | 0.12 |
ENST00000285311.3 |
DKK2 |
dickkopf WNT signaling pathway inhibitor 2 |
| chr1_+_210001309 | 0.12 |
ENST00000491415.2 |
DIEXF |
digestive organ expansion factor homolog (zebrafish) |
| chr5_+_95066823 | 0.12 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr12_-_120765565 | 0.12 |
ENST00000423423.3 ENST00000308366.4 |
PLA2G1B |
phospholipase A2, group IB (pancreas) |
| chr2_-_163008903 | 0.12 |
ENST00000418842.2 ENST00000375497.3 |
GCG |
glucagon |
| chr9_+_71939488 | 0.12 |
ENST00000455972.1 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr13_-_26625169 | 0.12 |
ENST00000319420.3 |
SHISA2 |
shisa family member 2 |
| chr7_+_107224364 | 0.12 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr15_-_89755034 | 0.12 |
ENST00000563254.1 |
RLBP1 |
retinaldehyde binding protein 1 |
| chr11_-_89654935 | 0.12 |
ENST00000530311.2 |
TRIM49D1 |
tripartite motif containing 49D1 |
| chr13_-_47471155 | 0.12 |
ENST00000543956.1 ENST00000542664.1 |
HTR2A |
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr3_-_194373831 | 0.12 |
ENST00000437613.1 |
LSG1 |
large 60S subunit nuclear export GTPase 1 |
| chr12_-_12849073 | 0.12 |
ENST00000332427.2 ENST00000540796.1 |
GPR19 |
G protein-coupled receptor 19 |
| chr12_-_125473600 | 0.12 |
ENST00000308736.2 |
DHX37 |
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
| chr5_-_180665195 | 0.12 |
ENST00000509148.1 |
GNB2L1 |
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr9_-_110540419 | 0.11 |
ENST00000398726.3 |
AL162389.1 |
Uncharacterized protein |
| chr2_+_170335924 | 0.11 |
ENST00000554017.1 ENST00000392663.2 ENST00000513963.1 |
BBS5 RP11-724O16.1 |
Bardet-Biedl syndrome 5 Bardet-Biedl syndrome 5 protein; Uncharacterized protein |
| chr14_+_73634537 | 0.11 |
ENST00000406768.1 |
PSEN1 |
presenilin 1 |
| chr14_-_53619816 | 0.11 |
ENST00000323669.5 ENST00000395606.1 ENST00000357758.3 |
DDHD1 |
DDHD domain containing 1 |
| chr22_+_21996549 | 0.11 |
ENST00000248958.4 |
SDF2L1 |
stromal cell-derived factor 2-like 1 |
| chr16_+_31366536 | 0.11 |
ENST00000562522.1 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr12_-_13248598 | 0.11 |
ENST00000337630.6 ENST00000545699.1 |
GSG1 |
germ cell associated 1 |
| chr17_+_55173933 | 0.11 |
ENST00000539273.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr3_-_65583561 | 0.11 |
ENST00000460329.2 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr17_-_19648916 | 0.11 |
ENST00000444455.1 ENST00000439102.2 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr3_-_160823040 | 0.11 |
ENST00000484127.1 ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr3_-_160823158 | 0.11 |
ENST00000392779.2 ENST00000392780.1 ENST00000494173.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr6_-_56707943 | 0.11 |
ENST00000370769.4 ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST |
dystonin |
| chr8_+_54764346 | 0.11 |
ENST00000297313.3 ENST00000344277.6 |
RGS20 |
regulator of G-protein signaling 20 |
| chrX_+_139791917 | 0.11 |
ENST00000607004.1 ENST00000370535.3 |
LINC00632 |
long intergenic non-protein coding RNA 632 |
| chr2_+_97001491 | 0.11 |
ENST00000240423.4 ENST00000427946.1 ENST00000435975.1 ENST00000456906.1 ENST00000455200.1 |
NCAPH |
non-SMC condensin I complex, subunit H |
| chr8_-_87081926 | 0.11 |
ENST00000276616.2 |
PSKH2 |
protein serine kinase H2 |
| chr14_+_105957402 | 0.11 |
ENST00000421892.1 ENST00000334656.7 ENST00000451719.1 ENST00000392522.3 ENST00000392523.4 ENST00000354560.6 ENST00000450383.1 |
C14orf80 |
chromosome 14 open reading frame 80 |
| chr12_-_96794143 | 0.11 |
ENST00000543119.2 |
CDK17 |
cyclin-dependent kinase 17 |
| chr11_+_93394805 | 0.11 |
ENST00000325212.6 ENST00000411936.1 ENST00000344196.4 |
KIAA1731 |
KIAA1731 |
| chr7_+_135242652 | 0.11 |
ENST00000285968.6 ENST00000440390.2 |
NUP205 |
nucleoporin 205kDa |
| chr10_-_103578182 | 0.11 |
ENST00000439817.1 |
MGEA5 |
meningioma expressed antigen 5 (hyaluronidase) |
| chr9_-_131486367 | 0.11 |
ENST00000372663.4 ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12 |
zinc finger, DHHC-type containing 12 |
| chr11_-_116663127 | 0.11 |
ENST00000433069.1 ENST00000542499.1 |
APOA5 |
apolipoprotein A-V |
| chr18_+_8717369 | 0.11 |
ENST00000359865.3 ENST00000400050.3 ENST00000306285.7 |
SOGA2 |
SOGA family member 2 |
| chr16_+_3068393 | 0.11 |
ENST00000573001.1 |
TNFRSF12A |
tumor necrosis factor receptor superfamily, member 12A |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1904170 | regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.2 | 0.5 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.1 | 0.5 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.3 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 0.1 | 0.4 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.2 | GO:0072186 | metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.2 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.2 | GO:0031938 | regulation of chromatin silencing at telomere(GO:0031938) |
| 0.1 | 0.2 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 0.2 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.1 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.5 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.1 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.2 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.2 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.2 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.0 | 0.2 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0035711 | T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.1 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0070428 | negative regulation of interleukin-23 production(GO:0032707) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.3 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.2 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.0 | 0.3 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.0 | GO:1902908 | regulation of melanosome transport(GO:1902908) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:1904729 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.6 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.1 | GO:0034699 | response to luteinizing hormone(GO:0034699) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.0 | GO:0015827 | tryptophan transport(GO:0015827) |
| 0.0 | 0.0 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.3 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 0.1 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.0 | 0.1 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.0 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.2 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0070982 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.0 | 0.0 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.0 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0072711 | cellular response to hydroxyurea(GO:0072711) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.0 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0045337 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.1 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.0 | 0.0 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.0 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.0 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0043318 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.0 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.0 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.0 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0051643 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) secretory granule localization(GO:0032252) endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.0 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.0 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.1 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.5 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.0 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.0 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.0 | GO:0018011 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.0 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.0 | GO:0033561 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.0 | 0.1 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.0 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.0 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.0 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.0 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.0 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.8 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.3 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.2 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.2 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0097181 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.2 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.0 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.0 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.3 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.2 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.2 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 0.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.0 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.0 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.0 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.0 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.0 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.0 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.0 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.0 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.0 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.0 | ST GAQ PATHWAY | G alpha q Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.7 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |