Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
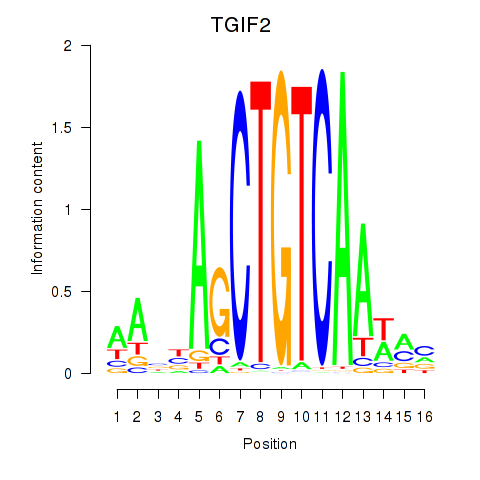
Results for PKNOX1_TGIF2
Z-value: 0.16
Transcription factors associated with PKNOX1_TGIF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX1
|
ENSG00000160199.10 | PKNOX1 |
|
TGIF2
|
ENSG00000118707.5 | TGIF2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PKNOX1 | hg19_v2_chr21_+_44394742_44394756 | 0.69 | 5.9e-02 | Click! |
| TGIF2 | hg19_v2_chr20_+_35201857_35201891, hg19_v2_chr20_+_35201993_35202050 | 0.32 | 4.3e-01 | Click! |
Activity profile of PKNOX1_TGIF2 motif
Sorted Z-values of PKNOX1_TGIF2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX1_TGIF2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_233925064 | 0.23 |
ENST00000359570.5 ENST00000538935.1 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
| chr19_-_38743878 | 0.22 |
ENST00000587515.1 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr1_+_145549203 | 0.22 |
ENST00000355594.4 ENST00000544626.1 |
ANKRD35 |
ankyrin repeat domain 35 |
| chrX_-_39923656 | 0.22 |
ENST00000413905.1 |
BCOR |
BCL6 corepressor |
| chr8_-_8318847 | 0.21 |
ENST00000521218.1 |
CTA-398F10.2 |
CTA-398F10.2 |
| chr2_-_203103185 | 0.20 |
ENST00000409205.1 |
SUMO1 |
small ubiquitin-like modifier 1 |
| chr3_-_169587621 | 0.20 |
ENST00000523069.1 ENST00000316428.5 ENST00000264676.5 |
LRRC31 |
leucine rich repeat containing 31 |
| chr9_-_130635741 | 0.20 |
ENST00000223836.10 |
AK1 |
adenylate kinase 1 |
| chr1_+_207262170 | 0.19 |
ENST00000367078.3 |
C4BPB |
complement component 4 binding protein, beta |
| chrX_+_101380642 | 0.18 |
ENST00000372780.1 ENST00000329035.2 |
TCEAL2 |
transcription elongation factor A (SII)-like 2 |
| chr1_+_207262578 | 0.18 |
ENST00000243611.5 ENST00000367076.3 |
C4BPB |
complement component 4 binding protein, beta |
| chr22_+_39052632 | 0.18 |
ENST00000411557.1 ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1 |
chibby homolog 1 (Drosophila) |
| chr1_+_207262627 | 0.18 |
ENST00000391923.1 |
C4BPB |
complement component 4 binding protein, beta |
| chr11_+_8932828 | 0.17 |
ENST00000530281.1 ENST00000396648.2 ENST00000534147.1 ENST00000529942.1 |
AKIP1 |
A kinase (PRKA) interacting protein 1 |
| chr1_+_28844648 | 0.17 |
ENST00000373832.1 ENST00000373831.3 |
RCC1 |
regulator of chromosome condensation 1 |
| chr1_-_51425902 | 0.17 |
ENST00000396153.2 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
| chr1_+_207262540 | 0.17 |
ENST00000452902.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr21_-_34185944 | 0.17 |
ENST00000479548.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr1_-_54872059 | 0.16 |
ENST00000371320.3 |
SSBP3 |
single stranded DNA binding protein 3 |
| chr18_-_52626622 | 0.16 |
ENST00000591504.1 |
CCDC68 |
coiled-coil domain containing 68 |
| chr16_-_33647696 | 0.16 |
ENST00000558425.1 ENST00000569103.2 |
RP11-812E19.9 |
Uncharacterized protein |
| chr3_-_190167571 | 0.16 |
ENST00000354905.2 |
TMEM207 |
transmembrane protein 207 |
| chr6_-_32140886 | 0.16 |
ENST00000395496.1 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr17_+_39405939 | 0.15 |
ENST00000334109.2 |
KRTAP9-4 |
keratin associated protein 9-4 |
| chr12_-_31882108 | 0.15 |
ENST00000281471.6 |
AMN1 |
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr17_-_40337470 | 0.15 |
ENST00000293330.1 |
HCRT |
hypocretin (orexin) neuropeptide precursor |
| chr6_-_132967142 | 0.14 |
ENST00000275216.1 |
TAAR1 |
trace amine associated receptor 1 |
| chr17_+_38296576 | 0.14 |
ENST00000264645.7 |
CASC3 |
cancer susceptibility candidate 3 |
| chr16_+_618837 | 0.14 |
ENST00000409439.2 |
PIGQ |
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr17_-_79827808 | 0.14 |
ENST00000580685.1 |
ARHGDIA |
Rho GDP dissociation inhibitor (GDI) alpha |
| chr4_-_114682364 | 0.14 |
ENST00000511664.1 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr1_-_205391178 | 0.13 |
ENST00000367153.4 ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1 |
LEM domain containing 1 |
| chr6_+_37225540 | 0.13 |
ENST00000373491.3 |
TBC1D22B |
TBC1 domain family, member 22B |
| chr3_+_111260954 | 0.13 |
ENST00000283285.5 |
CD96 |
CD96 molecule |
| chr2_+_113885138 | 0.13 |
ENST00000409930.3 |
IL1RN |
interleukin 1 receptor antagonist |
| chr3_+_111260856 | 0.13 |
ENST00000352690.4 |
CD96 |
CD96 molecule |
| chr7_-_132261223 | 0.13 |
ENST00000423507.2 |
PLXNA4 |
plexin A4 |
| chr2_+_10560147 | 0.13 |
ENST00000422133.1 |
HPCAL1 |
hippocalcin-like 1 |
| chr11_-_101000445 | 0.12 |
ENST00000534013.1 |
PGR |
progesterone receptor |
| chrX_-_71792477 | 0.12 |
ENST00000421523.1 ENST00000415409.1 ENST00000373559.4 ENST00000373556.3 ENST00000373560.2 ENST00000373583.1 ENST00000429103.2 ENST00000373571.1 ENST00000373554.1 |
HDAC8 |
histone deacetylase 8 |
| chr4_-_114682224 | 0.12 |
ENST00000342666.5 ENST00000515496.1 ENST00000514328.1 ENST00000508738.1 ENST00000379773.2 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr1_+_154966058 | 0.12 |
ENST00000392487.1 |
LENEP |
lens epithelial protein |
| chr2_-_3504587 | 0.12 |
ENST00000415131.1 |
ADI1 |
acireductone dioxygenase 1 |
| chr15_+_25068773 | 0.12 |
ENST00000400100.1 ENST00000400098.1 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr2_+_234959376 | 0.12 |
ENST00000425558.1 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
| chr14_-_21994525 | 0.12 |
ENST00000538754.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr14_+_100842735 | 0.12 |
ENST00000554998.1 ENST00000402312.3 ENST00000335290.6 ENST00000554175.1 |
WDR25 |
WD repeat domain 25 |
| chr19_+_9296279 | 0.11 |
ENST00000344248.2 |
OR7D2 |
olfactory receptor, family 7, subfamily D, member 2 |
| chr9_-_132805430 | 0.11 |
ENST00000446176.2 ENST00000355681.3 ENST00000420781.1 |
FNBP1 |
formin binding protein 1 |
| chr6_-_35656685 | 0.11 |
ENST00000539068.1 ENST00000540787.1 |
FKBP5 |
FK506 binding protein 5 |
| chr2_-_203103281 | 0.11 |
ENST00000392244.3 ENST00000409181.1 ENST00000409712.1 ENST00000409498.2 ENST00000409368.1 ENST00000392245.1 ENST00000392246.2 |
SUMO1 |
small ubiquitin-like modifier 1 |
| chrX_-_101397433 | 0.11 |
ENST00000372774.3 |
TCEAL6 |
transcription elongation factor A (SII)-like 6 |
| chr2_-_211179883 | 0.10 |
ENST00000352451.3 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr3_+_52719936 | 0.10 |
ENST00000418458.1 ENST00000394799.2 |
GNL3 |
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr6_+_54172653 | 0.10 |
ENST00000370869.3 |
TINAG |
tubulointerstitial nephritis antigen |
| chr11_-_71810258 | 0.10 |
ENST00000544594.1 |
LAMTOR1 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr18_+_3449821 | 0.10 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr1_+_207262881 | 0.10 |
ENST00000451804.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr12_+_69201923 | 0.10 |
ENST00000462284.1 ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2 |
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr21_+_38792602 | 0.10 |
ENST00000398960.2 ENST00000398956.2 |
DYRK1A |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr18_+_3451584 | 0.10 |
ENST00000551541.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr7_-_132261253 | 0.10 |
ENST00000321063.4 |
PLXNA4 |
plexin A4 |
| chr11_-_64052111 | 0.10 |
ENST00000394532.3 ENST00000394531.3 ENST00000309032.3 |
BAD |
BCL2-associated agonist of cell death |
| chr11_+_129245796 | 0.09 |
ENST00000281437.4 |
BARX2 |
BARX homeobox 2 |
| chr7_+_129015484 | 0.09 |
ENST00000490911.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr5_+_43603229 | 0.09 |
ENST00000344920.4 ENST00000512996.2 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr19_-_39322299 | 0.09 |
ENST00000601094.1 ENST00000595567.1 ENST00000602115.1 ENST00000601778.1 ENST00000597205.1 ENST00000595470.1 |
ECH1 |
enoyl CoA hydratase 1, peroxisomal |
| chrX_-_153775426 | 0.09 |
ENST00000393562.2 |
G6PD |
glucose-6-phosphate dehydrogenase |
| chr3_+_57875711 | 0.09 |
ENST00000442599.2 |
SLMAP |
sarcolemma associated protein |
| chr12_+_72148614 | 0.09 |
ENST00000261263.3 |
RAB21 |
RAB21, member RAS oncogene family |
| chr11_+_55578854 | 0.09 |
ENST00000333973.2 |
OR5L1 |
olfactory receptor, family 5, subfamily L, member 1 |
| chr12_+_55248289 | 0.09 |
ENST00000308796.6 |
MUCL1 |
mucin-like 1 |
| chr3_+_52828805 | 0.09 |
ENST00000416872.2 ENST00000449956.2 |
ITIH3 |
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr4_+_146539415 | 0.09 |
ENST00000281317.5 |
MMAA |
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr5_-_173043591 | 0.09 |
ENST00000285908.5 ENST00000480951.1 ENST00000311086.4 |
BOD1 |
biorientation of chromosomes in cell division 1 |
| chr8_+_97506033 | 0.09 |
ENST00000518385.1 |
SDC2 |
syndecan 2 |
| chr17_+_6916957 | 0.08 |
ENST00000547302.2 |
RNASEK-C17orf49 |
RNASEK-C17orf49 readthrough |
| chr10_-_79398127 | 0.08 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr2_+_191208196 | 0.08 |
ENST00000392329.2 ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1 |
inositol polyphosphate-1-phosphatase |
| chr8_-_141774467 | 0.08 |
ENST00000520151.1 ENST00000519024.1 ENST00000519465.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr2_+_113033164 | 0.08 |
ENST00000409871.1 ENST00000343936.4 |
ZC3H6 |
zinc finger CCCH-type containing 6 |
| chr3_-_142166904 | 0.08 |
ENST00000264951.4 |
XRN1 |
5'-3' exoribonuclease 1 |
| chr22_-_37172111 | 0.08 |
ENST00000417951.2 ENST00000430701.1 ENST00000433985.2 |
IFT27 |
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr22_-_37172191 | 0.08 |
ENST00000340630.5 |
IFT27 |
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr17_-_1420006 | 0.08 |
ENST00000320345.6 ENST00000406424.4 |
INPP5K |
inositol polyphosphate-5-phosphatase K |
| chr10_+_94608245 | 0.08 |
ENST00000443748.2 ENST00000260762.6 |
EXOC6 |
exocyst complex component 6 |
| chr1_+_3668962 | 0.08 |
ENST00000294600.2 |
CCDC27 |
coiled-coil domain containing 27 |
| chr2_+_234668894 | 0.08 |
ENST00000305208.5 ENST00000608383.1 ENST00000360418.3 |
UGT1A8 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr11_+_125616184 | 0.08 |
ENST00000305738.5 ENST00000437148.2 |
PATE1 |
prostate and testis expressed 1 |
| chr12_+_121647868 | 0.08 |
ENST00000359949.7 ENST00000541532.1 ENST00000543171.1 ENST00000538701.1 |
P2RX4 |
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr11_-_2924720 | 0.07 |
ENST00000455942.2 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr18_+_3451646 | 0.07 |
ENST00000345133.5 ENST00000330513.5 ENST00000549546.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr17_-_26220366 | 0.07 |
ENST00000460380.2 ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9 RP1-66C13.4 |
LYR motif containing 9 Uncharacterized protein |
| chr9_-_100459639 | 0.07 |
ENST00000375128.4 |
XPA |
xeroderma pigmentosum, complementation group A |
| chr8_+_11666649 | 0.07 |
ENST00000528643.1 ENST00000525777.1 |
FDFT1 |
farnesyl-diphosphate farnesyltransferase 1 |
| chr5_-_81046904 | 0.07 |
ENST00000515395.1 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr14_-_21493123 | 0.07 |
ENST00000556147.1 ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2 |
NDRG family member 2 |
| chr14_-_100842588 | 0.07 |
ENST00000556645.1 ENST00000556209.1 ENST00000556504.1 ENST00000556435.1 ENST00000554772.1 ENST00000553581.1 ENST00000553769.2 ENST00000554605.1 ENST00000557722.1 ENST00000553413.1 ENST00000553524.1 ENST00000358655.4 |
WARS |
tryptophanyl-tRNA synthetase |
| chr12_+_10658489 | 0.07 |
ENST00000538173.1 |
EIF2S3L |
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr3_+_38017264 | 0.07 |
ENST00000436654.1 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr10_+_28822636 | 0.06 |
ENST00000442148.1 ENST00000448193.1 |
WAC |
WW domain containing adaptor with coiled-coil |
| chr2_+_16080659 | 0.06 |
ENST00000281043.3 |
MYCN |
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr4_-_175443484 | 0.06 |
ENST00000514584.1 ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr12_-_15114191 | 0.06 |
ENST00000541380.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr9_-_134615443 | 0.06 |
ENST00000372195.1 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr12_+_103981044 | 0.06 |
ENST00000388887.2 |
STAB2 |
stabilin 2 |
| chr8_+_97657449 | 0.06 |
ENST00000220763.5 |
CPQ |
carboxypeptidase Q |
| chr3_-_52719810 | 0.06 |
ENST00000424867.1 ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1 |
polybromo 1 |
| chr2_+_234959323 | 0.06 |
ENST00000373368.1 ENST00000168148.3 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
| chr12_+_4699244 | 0.06 |
ENST00000540757.2 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr12_-_21757774 | 0.06 |
ENST00000261195.2 |
GYS2 |
glycogen synthase 2 (liver) |
| chr17_-_1420182 | 0.06 |
ENST00000421807.2 |
INPP5K |
inositol polyphosphate-5-phosphatase K |
| chr10_+_114710516 | 0.06 |
ENST00000542695.1 ENST00000346198.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr8_-_99837856 | 0.06 |
ENST00000518165.1 ENST00000419617.2 |
STK3 |
serine/threonine kinase 3 |
| chr16_-_69418649 | 0.06 |
ENST00000566257.1 |
TERF2 |
telomeric repeat binding factor 2 |
| chr1_-_214638146 | 0.06 |
ENST00000543945.1 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
| chr1_+_74701062 | 0.06 |
ENST00000326637.3 |
TNNI3K |
TNNI3 interacting kinase |
| chr7_-_104909435 | 0.06 |
ENST00000357311.3 |
SRPK2 |
SRSF protein kinase 2 |
| chr18_+_48918368 | 0.06 |
ENST00000583982.1 ENST00000578152.1 ENST00000583609.1 ENST00000435144.1 ENST00000580841.1 |
RP11-267C16.1 |
RP11-267C16.1 |
| chr5_-_81046841 | 0.05 |
ENST00000509013.2 ENST00000505980.1 ENST00000509053.1 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr12_+_122326662 | 0.05 |
ENST00000261817.2 ENST00000538613.1 ENST00000542602.1 |
PSMD9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr12_-_105478339 | 0.05 |
ENST00000424857.2 ENST00000258494.9 |
ALDH1L2 |
aldehyde dehydrogenase 1 family, member L2 |
| chr18_+_3449330 | 0.05 |
ENST00000549253.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr11_-_842509 | 0.05 |
ENST00000322028.4 |
POLR2L |
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr4_-_175443943 | 0.05 |
ENST00000296522.6 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr6_-_29324054 | 0.05 |
ENST00000543825.1 |
OR5V1 |
olfactory receptor, family 5, subfamily V, member 1 |
| chr20_+_54987305 | 0.05 |
ENST00000371336.3 ENST00000434344.1 |
CASS4 |
Cas scaffolding protein family member 4 |
| chr14_-_106573756 | 0.05 |
ENST00000390601.2 |
IGHV3-11 |
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr1_+_24285599 | 0.05 |
ENST00000471915.1 |
PNRC2 |
proline-rich nuclear receptor coactivator 2 |
| chr20_-_33872518 | 0.05 |
ENST00000374436.3 |
EIF6 |
eukaryotic translation initiation factor 6 |
| chr3_-_111852128 | 0.05 |
ENST00000308910.4 |
GCSAM |
germinal center-associated, signaling and motility |
| chr4_-_140477928 | 0.05 |
ENST00000274031.3 |
SETD7 |
SET domain containing (lysine methyltransferase) 7 |
| chr15_-_77197620 | 0.05 |
ENST00000565970.1 ENST00000563290.1 ENST00000565372.1 ENST00000564177.1 ENST00000568382.1 ENST00000563919.1 |
SCAPER |
S-phase cyclin A-associated protein in the ER |
| chr11_+_120207787 | 0.05 |
ENST00000397843.2 ENST00000356641.3 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr1_+_205473720 | 0.05 |
ENST00000429964.2 ENST00000506784.1 ENST00000360066.2 |
CDK18 |
cyclin-dependent kinase 18 |
| chr4_-_47983519 | 0.05 |
ENST00000358519.4 ENST00000544810.1 ENST00000402813.3 |
CNGA1 |
cyclic nucleotide gated channel alpha 1 |
| chr19_-_39322497 | 0.05 |
ENST00000221418.4 |
ECH1 |
enoyl CoA hydratase 1, peroxisomal |
| chr5_-_179051579 | 0.05 |
ENST00000505811.1 ENST00000515714.1 ENST00000513225.1 ENST00000503664.1 ENST00000356731.5 ENST00000523137.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr20_-_33872548 | 0.05 |
ENST00000374443.3 |
EIF6 |
eukaryotic translation initiation factor 6 |
| chr20_+_54987168 | 0.05 |
ENST00000360314.3 |
CASS4 |
Cas scaffolding protein family member 4 |
| chr3_-_142166846 | 0.05 |
ENST00000463916.1 ENST00000544157.1 |
XRN1 |
5'-3' exoribonuclease 1 |
| chr16_+_20817839 | 0.05 |
ENST00000348433.6 ENST00000568501.1 ENST00000566276.1 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr5_-_36301984 | 0.05 |
ENST00000502994.1 ENST00000515759.1 ENST00000296604.3 |
RANBP3L |
RAN binding protein 3-like |
| chr16_+_20818020 | 0.05 |
ENST00000564274.1 ENST00000563068.1 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr20_+_42187682 | 0.05 |
ENST00000373092.3 ENST00000373077.1 |
SGK2 |
serum/glucocorticoid regulated kinase 2 |
| chr7_+_76139741 | 0.05 |
ENST00000334348.3 ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B |
uroplakin 3B |
| chr20_+_42187608 | 0.05 |
ENST00000373100.1 |
SGK2 |
serum/glucocorticoid regulated kinase 2 |
| chr6_-_131949200 | 0.05 |
ENST00000539158.1 ENST00000368058.1 |
MED23 |
mediator complex subunit 23 |
| chr6_-_114664180 | 0.04 |
ENST00000312719.5 |
HS3ST5 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
| chr22_-_50219548 | 0.04 |
ENST00000404034.1 |
BRD1 |
bromodomain containing 1 |
| chr11_-_6440283 | 0.04 |
ENST00000299402.6 ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr21_+_30502806 | 0.04 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr9_-_123639600 | 0.04 |
ENST00000373896.3 |
PHF19 |
PHD finger protein 19 |
| chr19_+_36393422 | 0.04 |
ENST00000437550.2 |
HCST |
hematopoietic cell signal transducer |
| chr9_-_86432547 | 0.04 |
ENST00000376365.3 ENST00000376371.2 |
GKAP1 |
G kinase anchoring protein 1 |
| chr3_-_52569023 | 0.04 |
ENST00000307076.4 |
NT5DC2 |
5'-nucleotidase domain containing 2 |
| chr16_+_81678957 | 0.04 |
ENST00000398040.4 |
CMIP |
c-Maf inducing protein |
| chr14_-_21492251 | 0.04 |
ENST00000554398.1 |
NDRG2 |
NDRG family member 2 |
| chr4_+_86396321 | 0.04 |
ENST00000503995.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr3_+_57875738 | 0.04 |
ENST00000417128.1 ENST00000438794.1 |
SLMAP |
sarcolemma associated protein |
| chr4_-_149365827 | 0.04 |
ENST00000344721.4 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
| chr14_-_21492113 | 0.04 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr16_+_85061367 | 0.04 |
ENST00000538274.1 ENST00000258180.3 |
KIAA0513 |
KIAA0513 |
| chr15_+_25101698 | 0.04 |
ENST00000400097.1 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr7_-_42276612 | 0.04 |
ENST00000395925.3 ENST00000437480.1 |
GLI3 |
GLI family zinc finger 3 |
| chr19_+_1000418 | 0.04 |
ENST00000234389.3 |
GRIN3B |
glutamate receptor, ionotropic, N-methyl-D-aspartate 3B |
| chr16_+_20817761 | 0.04 |
ENST00000568046.1 ENST00000261377.6 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr8_-_18541603 | 0.04 |
ENST00000428502.2 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr1_-_63988846 | 0.04 |
ENST00000283568.8 ENST00000371092.3 ENST00000271002.10 |
ITGB3BP |
integrin beta 3 binding protein (beta3-endonexin) |
| chr2_-_225811747 | 0.04 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr11_+_107462471 | 0.04 |
ENST00000600612.1 |
AP000889.3 |
HCG2032453; Uncharacterized protein; cDNA FLJ25337 fis, clone TST00714 |
| chr9_-_123638633 | 0.04 |
ENST00000456291.1 |
PHF19 |
PHD finger protein 19 |
| chr15_+_36871983 | 0.04 |
ENST00000437989.2 ENST00000569302.1 |
C15orf41 |
chromosome 15 open reading frame 41 |
| chr3_-_27764190 | 0.04 |
ENST00000537516.1 |
EOMES |
eomesodermin |
| chr15_+_49170083 | 0.04 |
ENST00000530028.2 |
EID1 |
EP300 interacting inhibitor of differentiation 1 |
| chr6_+_168841817 | 0.04 |
ENST00000356284.2 ENST00000354536.5 |
SMOC2 |
SPARC related modular calcium binding 2 |
| chr2_+_198380289 | 0.04 |
ENST00000233892.4 ENST00000409916.1 |
MOB4 |
MOB family member 4, phocein |
| chr19_-_13044494 | 0.04 |
ENST00000593021.1 ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA |
phenylalanyl-tRNA synthetase, alpha subunit |
| chr14_+_22265444 | 0.04 |
ENST00000390430.2 |
TRAV8-1 |
T cell receptor alpha variable 8-1 |
| chr6_-_31651817 | 0.04 |
ENST00000375863.3 ENST00000375860.2 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr3_+_12329397 | 0.04 |
ENST00000397015.2 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr19_+_49496705 | 0.04 |
ENST00000595090.1 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr4_+_169418255 | 0.04 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr17_-_66951474 | 0.03 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr4_+_123653807 | 0.03 |
ENST00000314218.3 ENST00000542236.1 |
BBS12 |
Bardet-Biedl syndrome 12 |
| chr5_-_81046922 | 0.03 |
ENST00000514493.1 ENST00000320672.4 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr15_+_69222909 | 0.03 |
ENST00000455873.3 |
NOX5 |
NADPH oxidase, EF-hand calcium binding domain 5 |
| chr17_+_8191815 | 0.03 |
ENST00000226105.6 ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF |
RAN guanine nucleotide release factor |
| chr3_-_27763803 | 0.03 |
ENST00000449599.1 |
EOMES |
eomesodermin |
| chr1_-_33502528 | 0.03 |
ENST00000354858.6 |
AK2 |
adenylate kinase 2 |
| chr6_-_86099898 | 0.03 |
ENST00000455071.1 |
RP11-30P6.6 |
RP11-30P6.6 |
| chr15_+_43477455 | 0.03 |
ENST00000300213.4 |
CCNDBP1 |
cyclin D-type binding-protein 1 |
| chr3_+_50284321 | 0.03 |
ENST00000451956.1 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr12_+_69004619 | 0.03 |
ENST00000250559.9 ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B |
RAP1B, member of RAS oncogene family |
| chr3_+_45636219 | 0.03 |
ENST00000273317.4 |
LIMD1 |
LIM domains containing 1 |
| chr19_-_6424783 | 0.03 |
ENST00000398148.3 |
KHSRP |
KH-type splicing regulatory protein |
| chr4_+_159593418 | 0.03 |
ENST00000507475.1 ENST00000307738.5 |
ETFDH |
electron-transferring-flavoprotein dehydrogenase |
| chr16_-_4896205 | 0.03 |
ENST00000589389.1 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr16_-_2260834 | 0.03 |
ENST00000562360.1 ENST00000566018.1 |
BRICD5 |
BRICHOS domain containing 5 |
| chr19_+_11877838 | 0.03 |
ENST00000357901.4 ENST00000454339.2 |
ZNF441 |
zinc finger protein 441 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:1903027 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.1 | 0.3 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.2 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.3 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.1 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.3 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.1 | GO:1903823 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.2 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.2 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.0 | GO:2000870 | positive regulation of female gonad development(GO:2000196) regulation of progesterone secretion(GO:2000870) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.3 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.0 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.0 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |