Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
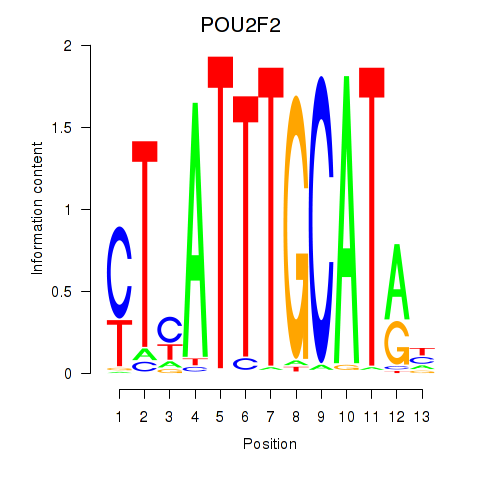
Results for POU2F2_POU3F1
Z-value: 0.63
Transcription factors associated with POU2F2_POU3F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU2F2
|
ENSG00000028277.16 | POU2F2 |
|
POU3F1
|
ENSG00000185668.5 | POU3F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F2 | hg19_v2_chr19_-_42636617_42636632 | 0.57 | 1.4e-01 | Click! |
| POU3F1 | hg19_v2_chr1_-_38512450_38512474 | -0.28 | 5.0e-01 | Click! |
Activity profile of POU2F2_POU3F1 motif
Sorted Z-values of POU2F2_POU3F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU2F2_POU3F1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_26124373 | 1.50 |
ENST00000377791.2 ENST00000602637.1 |
HIST1H2AC |
histone cluster 1, H2ac |
| chrX_-_24665208 | 1.26 |
ENST00000356768.4 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr5_+_36608422 | 1.11 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr9_-_140196703 | 0.88 |
ENST00000356628.2 |
NRARP |
NOTCH-regulated ankyrin repeat protein |
| chrX_-_24665353 | 0.73 |
ENST00000379144.2 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr18_-_53089723 | 0.72 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr12_+_113344755 | 0.65 |
ENST00000550883.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr6_+_37137939 | 0.65 |
ENST00000373509.5 |
PIM1 |
pim-1 oncogene |
| chr6_+_26251835 | 0.61 |
ENST00000356350.2 |
HIST1H2BH |
histone cluster 1, H2bh |
| chr12_-_31479045 | 0.61 |
ENST00000539409.1 ENST00000395766.1 |
FAM60A |
family with sequence similarity 60, member A |
| chr7_+_16793160 | 0.60 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr1_+_149822620 | 0.59 |
ENST00000369159.2 |
HIST2H2AA4 |
histone cluster 2, H2aa4 |
| chr17_-_47841485 | 0.59 |
ENST00000506156.1 ENST00000240364.2 |
FAM117A |
family with sequence similarity 117, member A |
| chr6_-_27114577 | 0.58 |
ENST00000356950.1 ENST00000396891.4 |
HIST1H2BK |
histone cluster 1, H2bk |
| chr6_-_26124138 | 0.58 |
ENST00000314332.5 ENST00000396984.1 |
HIST1H2BC |
histone cluster 1, H2bc |
| chr6_+_26183958 | 0.58 |
ENST00000356530.3 |
HIST1H2BE |
histone cluster 1, H2be |
| chr14_-_106622419 | 0.58 |
ENST00000390604.2 |
IGHV3-16 |
immunoglobulin heavy variable 3-16 (non-functional) |
| chr6_-_11779014 | 0.58 |
ENST00000229583.5 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr16_-_30107491 | 0.58 |
ENST00000566134.1 ENST00000565110.1 ENST00000398841.1 ENST00000398838.4 |
YPEL3 |
yippee-like 3 (Drosophila) |
| chr19_-_50143452 | 0.56 |
ENST00000246792.3 |
RRAS |
related RAS viral (r-ras) oncogene homolog |
| chr6_-_11779403 | 0.55 |
ENST00000414691.3 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr10_-_98031265 | 0.55 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr10_-_10836865 | 0.54 |
ENST00000446372.2 |
SFTA1P |
surfactant associated 1, pseudogene |
| chr6_-_27100529 | 0.54 |
ENST00000607124.1 ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ |
histone cluster 1, H2bj |
| chr14_-_106845789 | 0.53 |
ENST00000390617.2 |
IGHV3-35 |
immunoglobulin heavy variable 3-35 (non-functional) |
| chr22_+_32750872 | 0.53 |
ENST00000397468.1 |
RFPL3 |
ret finger protein-like 3 |
| chr10_-_10836919 | 0.52 |
ENST00000602763.1 ENST00000415590.2 ENST00000434919.2 |
SFTA1P |
surfactant associated 1, pseudogene |
| chr1_-_149814478 | 0.52 |
ENST00000369161.3 |
HIST2H2AA3 |
histone cluster 2, H2aa3 |
| chr1_-_149858227 | 0.50 |
ENST00000369155.2 |
HIST2H2BE |
histone cluster 2, H2be |
| chr16_-_33647696 | 0.49 |
ENST00000558425.1 ENST00000569103.2 |
RP11-812E19.9 |
Uncharacterized protein |
| chr19_+_17858509 | 0.49 |
ENST00000594202.1 ENST00000252771.7 ENST00000389133.4 |
FCHO1 |
FCH domain only 1 |
| chr10_-_98031310 | 0.49 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr9_-_124991124 | 0.46 |
ENST00000394319.4 ENST00000340587.3 |
LHX6 |
LIM homeobox 6 |
| chr16_+_33020496 | 0.46 |
ENST00000565407.2 |
IGHV3OR16-8 |
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr12_-_31479107 | 0.45 |
ENST00000542983.1 |
FAM60A |
family with sequence similarity 60, member A |
| chr5_+_145317356 | 0.45 |
ENST00000511217.1 |
SH3RF2 |
SH3 domain containing ring finger 2 |
| chr14_-_106610852 | 0.45 |
ENST00000390603.2 |
IGHV3-15 |
immunoglobulin heavy variable 3-15 |
| chr16_+_23847267 | 0.45 |
ENST00000321728.7 |
PRKCB |
protein kinase C, beta |
| chr14_-_34420259 | 0.44 |
ENST00000250457.3 ENST00000547327.2 |
EGLN3 |
egl-9 family hypoxia-inducible factor 3 |
| chr1_+_206730484 | 0.43 |
ENST00000304534.8 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
| chr2_+_207804278 | 0.41 |
ENST00000272852.3 |
CPO |
carboxypeptidase O |
| chr22_-_27456361 | 0.41 |
ENST00000453934.1 |
CTA-992D9.6 |
CTA-992D9.6 |
| chr4_-_186733363 | 0.40 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr12_+_113344811 | 0.39 |
ENST00000551241.1 ENST00000553185.1 ENST00000550689.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_-_23886285 | 0.38 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr22_+_23046750 | 0.38 |
ENST00000390307.2 |
IGLV3-22 |
immunoglobulin lambda variable 3-22 (gene/pseudogene) |
| chr18_-_52989217 | 0.38 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr2_+_68962014 | 0.37 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr1_+_156119466 | 0.37 |
ENST00000414683.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr6_-_99797522 | 0.36 |
ENST00000389677.5 |
FAXC |
failed axon connections homolog (Drosophila) |
| chr9_-_124990680 | 0.36 |
ENST00000541397.2 ENST00000560485.1 |
LHX6 |
LIM homeobox 6 |
| chrX_+_69674943 | 0.36 |
ENST00000542398.1 |
DLG3 |
discs, large homolog 3 (Drosophila) |
| chr17_-_19015945 | 0.35 |
ENST00000573866.2 |
SNORD3D |
small nucleolar RNA, C/D box 3D |
| chr18_-_52989525 | 0.35 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr6_+_27215471 | 0.34 |
ENST00000421826.2 |
PRSS16 |
protease, serine, 16 (thymus) |
| chr6_+_27215494 | 0.34 |
ENST00000230582.3 |
PRSS16 |
protease, serine, 16 (thymus) |
| chr7_-_41742697 | 0.34 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr6_-_10415470 | 0.34 |
ENST00000379604.2 ENST00000379613.3 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr14_+_65007177 | 0.34 |
ENST00000247207.6 |
HSPA2 |
heat shock 70kDa protein 2 |
| chr19_-_1132207 | 0.33 |
ENST00000438103.2 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
| chr1_-_149783914 | 0.32 |
ENST00000369167.1 ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF |
histone cluster 2, H2bf |
| chrX_-_117119243 | 0.32 |
ENST00000539496.1 ENST00000469946.1 |
KLHL13 |
kelch-like family member 13 |
| chr14_-_107013465 | 0.31 |
ENST00000390625.2 |
IGHV3-49 |
immunoglobulin heavy variable 3-49 |
| chr8_+_56792355 | 0.31 |
ENST00000519728.1 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr8_+_56792377 | 0.30 |
ENST00000520220.2 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr6_-_11779840 | 0.28 |
ENST00000506810.1 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr1_+_156119798 | 0.28 |
ENST00000355014.2 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr7_+_18535786 | 0.28 |
ENST00000406072.1 |
HDAC9 |
histone deacetylase 9 |
| chr11_-_94964354 | 0.28 |
ENST00000536441.1 |
SESN3 |
sestrin 3 |
| chr12_-_57328187 | 0.27 |
ENST00000293502.1 |
SDR9C7 |
short chain dehydrogenase/reductase family 9C, member 7 |
| chr17_+_73521763 | 0.26 |
ENST00000167462.5 ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2 |
lethal giant larvae homolog 2 (Drosophila) |
| chr12_+_107712173 | 0.26 |
ENST00000280758.5 ENST00000420571.2 |
BTBD11 |
BTB (POZ) domain containing 11 |
| chr12_+_113344582 | 0.26 |
ENST00000202917.5 ENST00000445409.2 ENST00000452357.2 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr17_-_7307358 | 0.26 |
ENST00000576017.1 ENST00000302422.3 ENST00000535512.1 |
TMEM256 TMEM256-PLSCR3 |
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr3_+_151591422 | 0.26 |
ENST00000362032.5 |
SUCNR1 |
succinate receptor 1 |
| chr1_+_6052700 | 0.25 |
ENST00000378092.1 ENST00000445501.1 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr19_+_3572758 | 0.25 |
ENST00000416526.1 |
HMG20B |
high mobility group 20B |
| chr2_+_90108504 | 0.25 |
ENST00000390271.2 |
IGKV6D-41 |
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr10_+_106113515 | 0.25 |
ENST00000369704.3 ENST00000312902.5 |
CCDC147 |
coiled-coil domain containing 147 |
| chr17_-_56494713 | 0.25 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr6_+_26158343 | 0.25 |
ENST00000377777.4 ENST00000289316.2 |
HIST1H2BD |
histone cluster 1, H2bd |
| chr17_-_56494908 | 0.25 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr17_-_56494882 | 0.25 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chr11_+_64085560 | 0.25 |
ENST00000265462.4 ENST00000352435.4 ENST00000347941.4 |
PRDX5 |
peroxiredoxin 5 |
| chr18_-_61311485 | 0.24 |
ENST00000436264.1 ENST00000356424.6 ENST00000341074.5 |
SERPINB4 |
serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| chr6_+_27114861 | 0.24 |
ENST00000377459.1 |
HIST1H2AH |
histone cluster 1, H2ah |
| chr11_-_64085533 | 0.24 |
ENST00000544844.1 |
TRMT112 |
tRNA methyltransferase 11-2 homolog (S. cerevisiae) |
| chr2_+_90060377 | 0.24 |
ENST00000436451.2 |
IGKV6D-21 |
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr14_-_72458326 | 0.24 |
ENST00000542853.1 |
AC005477.1 |
AC005477.1 |
| chr6_-_10415218 | 0.23 |
ENST00000466073.1 ENST00000498450.1 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr19_+_17337406 | 0.23 |
ENST00000597836.1 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr6_+_26273144 | 0.23 |
ENST00000377733.2 |
HIST1H2BI |
histone cluster 1, H2bi |
| chr22_-_29107919 | 0.23 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr12_-_7245125 | 0.22 |
ENST00000542285.1 ENST00000540610.1 |
C1R |
complement component 1, r subcomponent |
| chr8_+_19796381 | 0.22 |
ENST00000524029.1 ENST00000522701.1 ENST00000311322.8 |
LPL |
lipoprotein lipase |
| chr1_-_94586651 | 0.22 |
ENST00000535735.1 ENST00000370225.3 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr17_+_19091325 | 0.22 |
ENST00000584923.1 |
SNORD3A |
small nucleolar RNA, C/D box 3A |
| chr3_+_50192499 | 0.22 |
ENST00000413852.1 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr3_-_165555200 | 0.22 |
ENST00000479451.1 ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE |
butyrylcholinesterase |
| chr3_+_50192537 | 0.22 |
ENST00000002829.3 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr19_+_17337007 | 0.22 |
ENST00000215061.4 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr19_+_17337473 | 0.21 |
ENST00000598068.1 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr17_-_46894576 | 0.21 |
ENST00000393382.3 |
TTLL6 |
tubulin tyrosine ligase-like family, member 6 |
| chr1_+_167298281 | 0.21 |
ENST00000367862.5 |
POU2F1 |
POU class 2 homeobox 1 |
| chr14_-_106552755 | 0.21 |
ENST00000390600.2 |
IGHV3-9 |
immunoglobulin heavy variable 3-9 |
| chr4_-_47983519 | 0.21 |
ENST00000358519.4 ENST00000544810.1 ENST00000402813.3 |
CNGA1 |
cyclic nucleotide gated channel alpha 1 |
| chr11_-_64084959 | 0.21 |
ENST00000535750.1 ENST00000535126.1 ENST00000539854.1 ENST00000308774.2 |
TRMT112 |
tRNA methyltransferase 11-2 homolog (S. cerevisiae) |
| chrX_+_38660682 | 0.21 |
ENST00000457894.1 |
MID1IP1 |
MID1 interacting protein 1 |
| chr18_-_74839891 | 0.21 |
ENST00000581878.1 |
MBP |
myelin basic protein |
| chr3_-_126327398 | 0.21 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr6_-_52628271 | 0.21 |
ENST00000493422.1 |
GSTA2 |
glutathione S-transferase alpha 2 |
| chr11_-_62369291 | 0.20 |
ENST00000278823.2 |
MTA2 |
metastasis associated 1 family, member 2 |
| chr19_+_17337027 | 0.20 |
ENST00000601529.1 ENST00000600232.1 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr2_-_89459813 | 0.19 |
ENST00000390256.2 |
IGKV6-21 |
immunoglobulin kappa variable 6-21 (non-functional) |
| chr2_+_90211643 | 0.19 |
ENST00000390277.2 |
IGKV3D-11 |
immunoglobulin kappa variable 3D-11 |
| chr1_+_81771806 | 0.19 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr9_-_116065551 | 0.18 |
ENST00000297894.5 |
RNF183 |
ring finger protein 183 |
| chr19_-_19281054 | 0.18 |
ENST00000424583.2 ENST00000410050.1 ENST00000409224.1 ENST00000409447.2 |
MEF2B |
myocyte enhancer factor 2B |
| chr1_+_17575584 | 0.18 |
ENST00000375460.3 |
PADI3 |
peptidyl arginine deiminase, type III |
| chr4_-_40517984 | 0.18 |
ENST00000381795.6 |
RBM47 |
RNA binding motif protein 47 |
| chr6_+_27861190 | 0.18 |
ENST00000303806.4 |
HIST1H2BO |
histone cluster 1, H2bo |
| chrX_+_38660704 | 0.17 |
ENST00000378474.3 |
MID1IP1 |
MID1 interacting protein 1 |
| chr12_-_11062161 | 0.17 |
ENST00000390677.2 |
TAS2R13 |
taste receptor, type 2, member 13 |
| chr1_-_153044083 | 0.17 |
ENST00000341611.2 |
SPRR2B |
small proline-rich protein 2B |
| chr6_-_82462425 | 0.17 |
ENST00000369754.3 ENST00000320172.6 ENST00000369756.3 |
FAM46A |
family with sequence similarity 46, member A |
| chr6_-_27775694 | 0.17 |
ENST00000377401.2 |
HIST1H2BL |
histone cluster 1, H2bl |
| chr2_+_202047596 | 0.17 |
ENST00000286186.6 ENST00000360132.3 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr10_+_24528108 | 0.17 |
ENST00000438429.1 |
KIAA1217 |
KIAA1217 |
| chr15_+_52155001 | 0.17 |
ENST00000544199.1 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr19_-_44123734 | 0.16 |
ENST00000598676.1 |
ZNF428 |
zinc finger protein 428 |
| chr7_+_16566449 | 0.16 |
ENST00000401542.2 |
LRRC72 |
leucine rich repeat containing 72 |
| chr9_+_136325089 | 0.16 |
ENST00000291722.7 ENST00000316948.4 ENST00000540581.1 |
CACFD1 |
calcium channel flower domain containing 1 |
| chr4_+_5527117 | 0.16 |
ENST00000505296.1 |
C4orf6 |
chromosome 4 open reading frame 6 |
| chr6_-_143266297 | 0.16 |
ENST00000367603.2 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
| chr15_+_65337708 | 0.16 |
ENST00000334287.2 |
SLC51B |
solute carrier family 51, beta subunit |
| chr7_-_141673573 | 0.16 |
ENST00000547270.1 |
TAS2R38 |
taste receptor, type 2, member 38 |
| chr6_+_27775899 | 0.16 |
ENST00000358739.3 |
HIST1H2AI |
histone cluster 1, H2ai |
| chr4_+_5526883 | 0.16 |
ENST00000195455.2 |
C4orf6 |
chromosome 4 open reading frame 6 |
| chr11_-_75917569 | 0.16 |
ENST00000322563.3 |
WNT11 |
wingless-type MMTV integration site family, member 11 |
| chr2_-_89327228 | 0.16 |
ENST00000483158.1 |
IGKV3-11 |
immunoglobulin kappa variable 3-11 |
| chr7_-_100183742 | 0.16 |
ENST00000310300.6 |
LRCH4 |
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr15_+_45406519 | 0.16 |
ENST00000323030.5 |
DUOXA2 |
dual oxidase maturation factor 2 |
| chr6_+_26217159 | 0.16 |
ENST00000303910.2 |
HIST1H2AE |
histone cluster 1, H2ae |
| chr3_+_132036207 | 0.16 |
ENST00000336375.5 ENST00000495911.1 |
ACPP |
acid phosphatase, prostate |
| chr12_-_95941987 | 0.15 |
ENST00000537435.2 |
USP44 |
ubiquitin specific peptidase 44 |
| chr3_-_129407535 | 0.15 |
ENST00000432054.2 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
| chr19_+_50169081 | 0.15 |
ENST00000246784.3 |
BCL2L12 |
BCL2-like 12 (proline rich) |
| chr19_-_40791302 | 0.15 |
ENST00000392038.2 ENST00000578123.1 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
| chr12_-_48963829 | 0.15 |
ENST00000301046.2 ENST00000549817.1 |
LALBA |
lactalbumin, alpha- |
| chr3_-_112565703 | 0.15 |
ENST00000488794.1 |
CD200R1L |
CD200 receptor 1-like |
| chr6_+_43139037 | 0.14 |
ENST00000265354.4 |
SRF |
serum response factor (c-fos serum response element-binding transcription factor) |
| chr19_+_35849362 | 0.14 |
ENST00000327809.4 |
FFAR3 |
free fatty acid receptor 3 |
| chr17_-_10707410 | 0.14 |
ENST00000581851.1 |
LINC00675 |
long intergenic non-protein coding RNA 675 |
| chr19_+_41305330 | 0.14 |
ENST00000593972.1 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
| chr22_+_23161491 | 0.14 |
ENST00000390316.2 |
IGLV3-9 |
immunoglobulin lambda variable 3-9 (gene/pseudogene) |
| chr2_-_8464760 | 0.14 |
ENST00000430192.1 |
LINC00299 |
long intergenic non-protein coding RNA 299 |
| chr1_-_154832316 | 0.13 |
ENST00000361147.4 |
KCNN3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr6_+_31588478 | 0.13 |
ENST00000376007.4 ENST00000376033.2 |
PRRC2A |
proline-rich coiled-coil 2A |
| chr12_+_81110684 | 0.13 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chrX_-_84634737 | 0.13 |
ENST00000262753.4 |
POF1B |
premature ovarian failure, 1B |
| chr16_+_56691606 | 0.13 |
ENST00000334350.6 |
MT1F |
metallothionein 1F |
| chr15_+_67841330 | 0.13 |
ENST00000354498.5 |
MAP2K5 |
mitogen-activated protein kinase kinase 5 |
| chr2_+_114163945 | 0.13 |
ENST00000453673.3 |
IGKV1OR2-108 |
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr19_-_50169064 | 0.13 |
ENST00000593337.1 ENST00000598808.1 ENST00000600453.1 ENST00000593818.1 ENST00000597198.1 ENST00000601809.1 ENST00000377139.3 |
IRF3 |
interferon regulatory factor 3 |
| chr5_-_36301984 | 0.13 |
ENST00000502994.1 ENST00000515759.1 ENST00000296604.3 |
RANBP3L |
RAN binding protein 3-like |
| chr18_-_53253112 | 0.13 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chr2_+_220144052 | 0.12 |
ENST00000425450.1 ENST00000392086.4 ENST00000421532.1 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr2_-_89247338 | 0.12 |
ENST00000496168.1 |
IGKV1-5 |
immunoglobulin kappa variable 1-5 |
| chr4_+_184826418 | 0.12 |
ENST00000308497.4 ENST00000438269.1 |
STOX2 |
storkhead box 2 |
| chr3_-_24207039 | 0.12 |
ENST00000280696.5 |
THRB |
thyroid hormone receptor, beta |
| chr2_+_90273679 | 0.12 |
ENST00000423080.2 |
IGKV3D-7 |
immunoglobulin kappa variable 3D-7 |
| chr6_+_27782788 | 0.12 |
ENST00000359465.4 |
HIST1H2BM |
histone cluster 1, H2bm |
| chr6_-_33679452 | 0.12 |
ENST00000374231.4 ENST00000607484.1 ENST00000374214.3 |
UQCC2 |
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr12_-_56359802 | 0.12 |
ENST00000548803.1 ENST00000449260.2 ENST00000550447.1 ENST00000547137.1 ENST00000546543.1 ENST00000550464.1 |
PMEL |
premelanosome protein |
| chr19_+_19144384 | 0.12 |
ENST00000392335.2 ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6 |
armadillo repeat containing 6 |
| chr4_-_25865159 | 0.12 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr14_-_106866934 | 0.12 |
ENST00000390618.2 |
IGHV3-38 |
immunoglobulin heavy variable 3-38 (non-functional) |
| chr2_+_220143989 | 0.12 |
ENST00000336576.5 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr11_+_112047087 | 0.12 |
ENST00000526088.1 ENST00000532593.1 ENST00000531169.1 |
BCO2 |
beta-carotene oxygenase 2 |
| chr19_-_50168962 | 0.12 |
ENST00000599223.1 ENST00000593922.1 ENST00000600022.1 ENST00000596765.1 ENST00000599144.1 ENST00000596822.1 ENST00000598108.1 ENST00000601373.1 ENST00000595034.1 ENST00000601291.1 |
IRF3 |
interferon regulatory factor 3 |
| chr6_+_148663729 | 0.12 |
ENST00000367467.3 |
SASH1 |
SAM and SH3 domain containing 1 |
| chr12_-_100656134 | 0.11 |
ENST00000548313.1 |
DEPDC4 |
DEP domain containing 4 |
| chr5_-_169626104 | 0.11 |
ENST00000520275.1 ENST00000506431.2 |
CTB-27N1.1 |
CTB-27N1.1 |
| chr2_+_16080659 | 0.11 |
ENST00000281043.3 |
MYCN |
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr19_+_35861831 | 0.11 |
ENST00000454971.1 |
GPR42 |
G protein-coupled receptor 42 (gene/pseudogene) |
| chr21_+_17566643 | 0.11 |
ENST00000419952.1 ENST00000445461.2 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr14_-_107199464 | 0.11 |
ENST00000433072.2 |
IGHV3-72 |
immunoglobulin heavy variable 3-72 |
| chr19_+_56111680 | 0.11 |
ENST00000301073.3 |
ZNF524 |
zinc finger protein 524 |
| chr1_+_149858461 | 0.11 |
ENST00000331380.2 |
HIST2H2AC |
histone cluster 2, H2ac |
| chr12_-_122238464 | 0.11 |
ENST00000546227.1 |
RHOF |
ras homolog family member F (in filopodia) |
| chr19_-_41903161 | 0.11 |
ENST00000602129.1 ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5 |
exosome component 5 |
| chr6_-_26216872 | 0.10 |
ENST00000244601.3 |
HIST1H2BG |
histone cluster 1, H2bg |
| chr1_+_61547894 | 0.10 |
ENST00000403491.3 |
NFIA |
nuclear factor I/A |
| chr18_+_44526786 | 0.10 |
ENST00000245121.5 ENST00000356157.7 |
KATNAL2 |
katanin p60 subunit A-like 2 |
| chr19_-_44124019 | 0.10 |
ENST00000300811.3 |
ZNF428 |
zinc finger protein 428 |
| chr12_+_19358228 | 0.10 |
ENST00000424268.1 ENST00000543806.1 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr12_+_43086018 | 0.10 |
ENST00000550177.1 |
RP11-25I15.3 |
RP11-25I15.3 |
| chr12_+_7052974 | 0.10 |
ENST00000544681.1 ENST00000537087.1 |
C12orf57 |
chromosome 12 open reading frame 57 |
| chr3_+_132036243 | 0.10 |
ENST00000475741.1 ENST00000351273.7 |
ACPP |
acid phosphatase, prostate |
| chr2_+_90259593 | 0.10 |
ENST00000471857.1 |
IGKV1D-8 |
immunoglobulin kappa variable 1D-8 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070666 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.2 | 2.0 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.2 | 1.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 0.5 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.7 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.6 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.1 | 0.3 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 2.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.5 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 0.9 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.2 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.8 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.3 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 0.3 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.1 | 0.2 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.7 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.4 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.1 | 0.2 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.0 | 0.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.1 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.3 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0071484 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.4 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.3 | GO:0071888 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) macrophage apoptotic process(GO:0071888) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 2.1 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 1.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0048867 | ganglion mother cell fate determination(GO:0007402) stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.0 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 1.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein glycosylation(GO:0060050) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.4 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.0 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 0.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 5.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.3 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 1.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.5 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 1.6 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.3 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.1 | 0.7 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.2 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.2 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.0 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |