Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
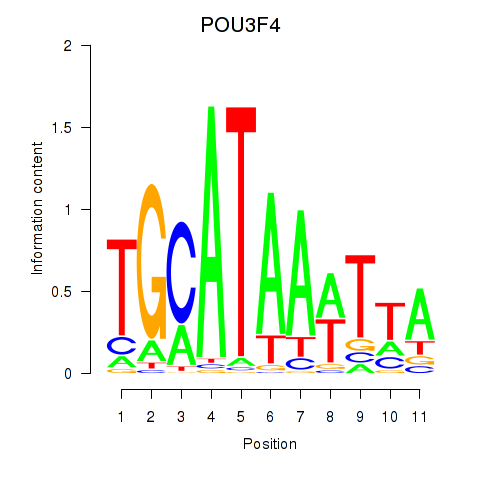
Results for POU3F3_POU3F4
Z-value: 0.42
Transcription factors associated with POU3F3_POU3F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU3F3
|
ENSG00000198914.2 | POU3F3 |
|
POU3F4
|
ENSG00000196767.4 | POU3F4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU3F4 | hg19_v2_chrX_+_82763265_82763283 | 0.54 | 1.7e-01 | Click! |
| POU3F3 | hg19_v2_chr2_+_105471969_105471969 | 0.06 | 8.8e-01 | Click! |
Activity profile of POU3F3_POU3F4 motif
Sorted Z-values of POU3F3_POU3F4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU3F3_POU3F4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_209602156 | 0.97 |
ENST00000429156.1 ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr1_+_209602609 | 0.87 |
ENST00000458250.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr10_+_71561649 | 0.45 |
ENST00000398978.3 ENST00000354547.3 ENST00000357811.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr8_+_31497271 | 0.43 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr10_+_71561630 | 0.35 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr3_-_112565703 | 0.35 |
ENST00000488794.1 |
CD200R1L |
CD200 receptor 1-like |
| chr20_+_58179582 | 0.34 |
ENST00000371015.1 ENST00000395639.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr3_+_189507432 | 0.32 |
ENST00000354600.5 |
TP63 |
tumor protein p63 |
| chr3_-_112564797 | 0.29 |
ENST00000398214.1 ENST00000448932.1 |
CD200R1L |
CD200 receptor 1-like |
| chrX_-_32173579 | 0.28 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr22_+_45148432 | 0.28 |
ENST00000389774.2 ENST00000396119.2 ENST00000336963.4 ENST00000356099.6 ENST00000412433.1 |
ARHGAP8 |
Rho GTPase activating protein 8 |
| chr8_+_61591337 | 0.26 |
ENST00000423902.2 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
| chrX_+_41548220 | 0.26 |
ENST00000378142.4 |
GPR34 |
G protein-coupled receptor 34 |
| chrX_+_41548259 | 0.25 |
ENST00000378138.5 |
GPR34 |
G protein-coupled receptor 34 |
| chr19_+_17858509 | 0.25 |
ENST00000594202.1 ENST00000252771.7 ENST00000389133.4 |
FCHO1 |
FCH domain only 1 |
| chr3_-_151034734 | 0.24 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr19_+_17858547 | 0.23 |
ENST00000600676.1 ENST00000600209.1 ENST00000596309.1 ENST00000598539.1 ENST00000597474.1 ENST00000593385.1 ENST00000598067.1 ENST00000593833.1 |
FCHO1 |
FCH domain only 1 |
| chr5_+_36608422 | 0.23 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr10_+_71561704 | 0.22 |
ENST00000520267.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr22_+_39101728 | 0.22 |
ENST00000216044.5 ENST00000484657.1 |
GTPBP1 |
GTP binding protein 1 |
| chr2_+_90060377 | 0.22 |
ENST00000436451.2 |
IGKV6D-21 |
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr10_+_24738355 | 0.21 |
ENST00000307544.6 |
KIAA1217 |
KIAA1217 |
| chr6_-_11779014 | 0.21 |
ENST00000229583.5 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr6_-_11779174 | 0.21 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr18_+_47088401 | 0.19 |
ENST00000261292.4 ENST00000427224.2 ENST00000580036.1 |
LIPG |
lipase, endothelial |
| chr6_-_11779403 | 0.19 |
ENST00000414691.3 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr8_-_41166953 | 0.19 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr6_-_136847099 | 0.19 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr10_-_116444371 | 0.18 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr12_+_41086297 | 0.18 |
ENST00000551295.2 |
CNTN1 |
contactin 1 |
| chr17_+_17685422 | 0.17 |
ENST00000395774.1 |
RAI1 |
retinoic acid induced 1 |
| chr10_-_75226166 | 0.17 |
ENST00000544628.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr17_-_39023462 | 0.15 |
ENST00000251643.4 |
KRT12 |
keratin 12 |
| chr1_+_101702417 | 0.15 |
ENST00000305352.6 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
| chr8_-_91095099 | 0.15 |
ENST00000265431.3 |
CALB1 |
calbindin 1, 28kDa |
| chr18_-_52989217 | 0.15 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr3_+_189507523 | 0.15 |
ENST00000437221.1 ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63 |
tumor protein p63 |
| chr1_+_186798073 | 0.14 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr14_-_21270561 | 0.14 |
ENST00000412779.2 |
RNASE1 |
ribonuclease, RNase A family, 1 (pancreatic) |
| chr17_-_3599696 | 0.14 |
ENST00000225328.5 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr6_-_9933500 | 0.14 |
ENST00000492169.1 |
OFCC1 |
orofacial cleft 1 candidate 1 |
| chr6_-_136847610 | 0.14 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr17_-_3599327 | 0.14 |
ENST00000551178.1 ENST00000552276.1 ENST00000547178.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr5_-_78809950 | 0.14 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr15_-_45670924 | 0.13 |
ENST00000396659.3 |
GATM |
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr1_+_62439037 | 0.13 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr10_+_695888 | 0.13 |
ENST00000441152.2 |
PRR26 |
proline rich 26 |
| chr18_-_52989525 | 0.13 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr19_-_14992264 | 0.12 |
ENST00000327462.2 |
OR7A17 |
olfactory receptor, family 7, subfamily A, member 17 |
| chr3_-_141747950 | 0.12 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr14_-_21270995 | 0.12 |
ENST00000555698.1 ENST00000397970.4 ENST00000340900.3 |
RNASE1 |
ribonuclease, RNase A family, 1 (pancreatic) |
| chr1_-_242612779 | 0.12 |
ENST00000427495.1 |
PLD5 |
phospholipase D family, member 5 |
| chr18_-_33647487 | 0.12 |
ENST00000590898.1 ENST00000357384.4 ENST00000319040.6 ENST00000588737.1 ENST00000399022.4 |
RPRD1A |
regulation of nuclear pre-mRNA domain containing 1A |
| chr5_+_36152179 | 0.12 |
ENST00000508514.1 ENST00000513151.1 ENST00000546211.1 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr8_+_31496809 | 0.12 |
ENST00000518104.1 ENST00000519301.1 |
NRG1 |
neuregulin 1 |
| chr8_-_38008783 | 0.11 |
ENST00000276449.4 |
STAR |
steroidogenic acute regulatory protein |
| chr4_-_100575781 | 0.11 |
ENST00000511828.1 |
RP11-766F14.2 |
Protein LOC285556 |
| chr3_-_151047327 | 0.10 |
ENST00000325602.5 |
P2RY13 |
purinergic receptor P2Y, G-protein coupled, 13 |
| chr8_-_49833978 | 0.10 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr18_+_616711 | 0.10 |
ENST00000579494.1 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr11_-_26593677 | 0.10 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr13_-_20110902 | 0.10 |
ENST00000390680.2 ENST00000382977.4 ENST00000382975.4 ENST00000457266.2 |
TPTE2 |
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr3_-_196911002 | 0.09 |
ENST00000452595.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr12_+_113354341 | 0.09 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr13_+_109248500 | 0.09 |
ENST00000356711.2 |
MYO16 |
myosin XVI |
| chr8_-_49834299 | 0.09 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr12_+_41221794 | 0.08 |
ENST00000547849.1 |
CNTN1 |
contactin 1 |
| chr15_-_65321943 | 0.08 |
ENST00000220058.4 |
MTFMT |
mitochondrial methionyl-tRNA formyltransferase |
| chrX_-_119445306 | 0.08 |
ENST00000371369.4 ENST00000440464.1 ENST00000519908.1 |
TMEM255A |
transmembrane protein 255A |
| chr4_-_87281224 | 0.08 |
ENST00000395169.3 ENST00000395161.2 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr5_+_36152091 | 0.08 |
ENST00000274254.5 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr3_-_141747439 | 0.08 |
ENST00000467667.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr10_+_85933494 | 0.08 |
ENST00000372126.3 |
C10orf99 |
chromosome 10 open reading frame 99 |
| chr19_+_17862274 | 0.08 |
ENST00000596536.1 ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1 |
FCH domain only 1 |
| chr8_-_124749609 | 0.08 |
ENST00000262219.6 ENST00000419625.1 |
ANXA13 |
annexin A13 |
| chr5_+_36152163 | 0.08 |
ENST00000274255.6 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr16_-_70835034 | 0.08 |
ENST00000261776.5 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
| chrX_+_130192318 | 0.08 |
ENST00000370922.1 |
ARHGAP36 |
Rho GTPase activating protein 36 |
| chr12_-_102874330 | 0.08 |
ENST00000307046.8 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr17_-_10452929 | 0.07 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr6_+_26273144 | 0.07 |
ENST00000377733.2 |
HIST1H2BI |
histone cluster 1, H2bi |
| chr6_-_76782371 | 0.07 |
ENST00000369950.3 ENST00000369963.3 |
IMPG1 |
interphotoreceptor matrix proteoglycan 1 |
| chr1_+_74701062 | 0.07 |
ENST00000326637.3 |
TNNI3K |
TNNI3 interacting kinase |
| chr7_-_36406750 | 0.07 |
ENST00000453212.1 ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895 |
KIAA0895 |
| chrX_-_131262048 | 0.07 |
ENST00000298542.4 |
FRMD7 |
FERM domain containing 7 |
| chr17_-_38821373 | 0.07 |
ENST00000394052.3 |
KRT222 |
keratin 222 |
| chr1_-_100231349 | 0.07 |
ENST00000287474.5 ENST00000414213.1 |
FRRS1 |
ferric-chelate reductase 1 |
| chr12_-_9760482 | 0.07 |
ENST00000229402.3 |
KLRB1 |
killer cell lectin-like receptor subfamily B, member 1 |
| chr14_+_70918874 | 0.07 |
ENST00000603540.1 |
ADAM21 |
ADAM metallopeptidase domain 21 |
| chr12_-_50616382 | 0.07 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr2_-_46769694 | 0.07 |
ENST00000522587.1 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr13_-_26795840 | 0.07 |
ENST00000381570.3 ENST00000399762.2 ENST00000346166.3 |
RNF6 |
ring finger protein (C3H2C3 type) 6 |
| chr17_+_17206635 | 0.07 |
ENST00000389022.4 |
NT5M |
5',3'-nucleotidase, mitochondrial |
| chr3_+_35722487 | 0.07 |
ENST00000441454.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr10_+_696000 | 0.07 |
ENST00000381489.5 |
PRR26 |
proline rich 26 |
| chrX_+_15525426 | 0.07 |
ENST00000342014.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chr10_+_29577974 | 0.06 |
ENST00000375500.3 |
LYZL1 |
lysozyme-like 1 |
| chr12_-_51611477 | 0.06 |
ENST00000389243.4 |
POU6F1 |
POU class 6 homeobox 1 |
| chr10_-_75676400 | 0.06 |
ENST00000412307.2 |
C10orf55 |
chromosome 10 open reading frame 55 |
| chr4_-_152149033 | 0.06 |
ENST00000514152.1 |
SH3D19 |
SH3 domain containing 19 |
| chr4_+_146403912 | 0.06 |
ENST00000507367.1 ENST00000394092.2 ENST00000515385.1 |
SMAD1 |
SMAD family member 1 |
| chr1_+_244515930 | 0.06 |
ENST00000366537.1 ENST00000308105.4 |
C1orf100 |
chromosome 1 open reading frame 100 |
| chr9_+_77230499 | 0.06 |
ENST00000396204.2 |
RORB |
RAR-related orphan receptor B |
| chr4_+_175839551 | 0.06 |
ENST00000404450.4 ENST00000514159.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chr1_+_6105974 | 0.06 |
ENST00000378083.3 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr12_+_21207503 | 0.06 |
ENST00000545916.1 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr1_-_216978709 | 0.06 |
ENST00000360012.3 |
ESRRG |
estrogen-related receptor gamma |
| chr21_+_17443521 | 0.06 |
ENST00000456342.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr17_-_41738931 | 0.06 |
ENST00000329168.3 ENST00000549132.1 |
MEOX1 |
mesenchyme homeobox 1 |
| chr7_-_105332084 | 0.06 |
ENST00000472195.1 |
ATXN7L1 |
ataxin 7-like 1 |
| chr1_-_193155729 | 0.05 |
ENST00000367434.4 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr17_-_41739283 | 0.05 |
ENST00000393661.2 ENST00000318579.4 |
MEOX1 |
mesenchyme homeobox 1 |
| chr11_-_104827425 | 0.05 |
ENST00000393150.3 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr13_-_46716969 | 0.05 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr15_+_52121822 | 0.05 |
ENST00000558455.1 ENST00000308580.7 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr21_+_17443434 | 0.05 |
ENST00000400178.2 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr4_-_168155700 | 0.05 |
ENST00000357545.4 ENST00000512648.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_+_30723003 | 0.05 |
ENST00000543491.1 |
PCDH7 |
protocadherin 7 |
| chr3_+_151591422 | 0.05 |
ENST00000362032.5 |
SUCNR1 |
succinate receptor 1 |
| chr7_+_119913688 | 0.05 |
ENST00000331113.4 |
KCND2 |
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr4_-_164395014 | 0.05 |
ENST00000280605.3 |
TKTL2 |
transketolase-like 2 |
| chr12_+_49297887 | 0.05 |
ENST00000266984.5 |
CCDC65 |
coiled-coil domain containing 65 |
| chr8_+_67039131 | 0.05 |
ENST00000315962.4 ENST00000353317.5 |
TRIM55 |
tripartite motif containing 55 |
| chr12_-_10324716 | 0.05 |
ENST00000545927.1 ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1 |
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr10_+_114710425 | 0.05 |
ENST00000352065.5 ENST00000369395.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_+_41086215 | 0.05 |
ENST00000547702.1 ENST00000551424.1 |
CNTN1 |
contactin 1 |
| chr14_-_65409502 | 0.05 |
ENST00000389614.5 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr8_-_70016408 | 0.05 |
ENST00000518540.1 |
RP11-600K15.1 |
RP11-600K15.1 |
| chr18_+_616672 | 0.05 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr14_+_22675388 | 0.04 |
ENST00000390461.2 |
TRAV34 |
T cell receptor alpha variable 34 |
| chr21_+_17442799 | 0.04 |
ENST00000602580.1 ENST00000458468.1 ENST00000602935.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr3_+_111805182 | 0.04 |
ENST00000430855.1 ENST00000431717.2 ENST00000264848.5 |
C3orf52 |
chromosome 3 open reading frame 52 |
| chr11_-_105010320 | 0.04 |
ENST00000532895.1 ENST00000530950.1 |
CARD18 |
caspase recruitment domain family, member 18 |
| chr3_-_193272874 | 0.04 |
ENST00000342695.4 |
ATP13A4 |
ATPase type 13A4 |
| chr14_-_65409438 | 0.04 |
ENST00000557049.1 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr1_+_73771844 | 0.04 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr4_-_168155730 | 0.04 |
ENST00000502330.1 ENST00000357154.3 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr19_-_44174330 | 0.04 |
ENST00000340093.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr6_-_66417107 | 0.04 |
ENST00000370621.3 ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS |
eyes shut homolog (Drosophila) |
| chr12_-_91451758 | 0.04 |
ENST00000266719.3 |
KERA |
keratocan |
| chr4_-_120243545 | 0.04 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr14_+_67291158 | 0.04 |
ENST00000555456.1 |
GPHN |
gephyrin |
| chr5_-_160973649 | 0.04 |
ENST00000393959.1 ENST00000517547.1 |
GABRB2 |
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr17_-_41050716 | 0.04 |
ENST00000417193.1 ENST00000301683.3 ENST00000436546.1 ENST00000431109.2 |
LINC00671 |
long intergenic non-protein coding RNA 671 |
| chrX_+_15518923 | 0.04 |
ENST00000348343.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chr3_+_173116225 | 0.04 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr16_+_31128978 | 0.04 |
ENST00000448516.2 ENST00000219797.4 |
KAT8 |
K(lysine) acetyltransferase 8 |
| chr7_-_16505440 | 0.04 |
ENST00000307068.4 |
SOSTDC1 |
sclerostin domain containing 1 |
| chr2_+_166095898 | 0.04 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chrX_+_102024075 | 0.04 |
ENST00000431616.1 ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630 |
long intergenic non-protein coding RNA 630 |
| chr1_+_167298281 | 0.04 |
ENST00000367862.5 |
POU2F1 |
POU class 2 homeobox 1 |
| chr2_-_113993020 | 0.04 |
ENST00000465084.1 |
PAX8 |
paired box 8 |
| chr2_-_228244013 | 0.04 |
ENST00000304568.3 |
TM4SF20 |
transmembrane 4 L six family member 20 |
| chr3_-_141747459 | 0.04 |
ENST00000477292.1 ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_-_102874102 | 0.04 |
ENST00000392905.2 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr10_+_5135981 | 0.04 |
ENST00000380554.3 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
| chr12_-_102224457 | 0.04 |
ENST00000549165.1 ENST00000549940.1 ENST00000392919.4 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chrX_+_130192216 | 0.04 |
ENST00000276211.5 |
ARHGAP36 |
Rho GTPase activating protein 36 |
| chr4_+_175839506 | 0.04 |
ENST00000505141.1 ENST00000359240.3 ENST00000445694.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chr5_-_11589131 | 0.04 |
ENST00000511377.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr2_+_28974668 | 0.04 |
ENST00000296122.6 ENST00000395366.2 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr15_-_76304731 | 0.04 |
ENST00000394907.3 |
NRG4 |
neuregulin 4 |
| chr12_+_25205446 | 0.04 |
ENST00000557489.1 ENST00000354454.3 ENST00000536173.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr20_+_16729003 | 0.03 |
ENST00000246081.2 |
OTOR |
otoraplin |
| chr2_+_145780725 | 0.03 |
ENST00000451478.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr4_-_168155577 | 0.03 |
ENST00000541354.1 ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr2_+_28974603 | 0.03 |
ENST00000441461.1 ENST00000358506.2 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
| chrX_-_77914825 | 0.03 |
ENST00000321110.1 |
ZCCHC5 |
zinc finger, CCHC domain containing 5 |
| chr9_-_95244781 | 0.03 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr2_+_120687335 | 0.03 |
ENST00000544261.1 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr12_-_39734783 | 0.03 |
ENST00000552961.1 |
KIF21A |
kinesin family member 21A |
| chr17_-_67057047 | 0.03 |
ENST00000495634.1 ENST00000453985.2 ENST00000585714.1 |
ABCA9 |
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr15_+_76352178 | 0.03 |
ENST00000388942.3 |
C15orf27 |
chromosome 15 open reading frame 27 |
| chr1_-_27701307 | 0.03 |
ENST00000270879.4 ENST00000354982.2 |
FCN3 |
ficolin (collagen/fibrinogen domain containing) 3 |
| chr21_-_27423339 | 0.03 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr2_+_89998789 | 0.03 |
ENST00000453166.2 |
IGKV2D-28 |
immunoglobulin kappa variable 2D-28 |
| chr19_+_36486078 | 0.03 |
ENST00000378887.2 |
SDHAF1 |
succinate dehydrogenase complex assembly factor 1 |
| chr4_-_110723335 | 0.03 |
ENST00000394634.2 |
CFI |
complement factor I |
| chr9_-_34458531 | 0.03 |
ENST00000379089.1 ENST00000379087.1 ENST00000379084.1 ENST00000379081.1 ENST00000379080.1 ENST00000422409.1 ENST00000379078.1 ENST00000445726.1 ENST00000297620.4 |
FAM219A |
family with sequence similarity 219, member A |
| chr4_-_110723134 | 0.03 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr16_+_75032901 | 0.03 |
ENST00000335325.4 ENST00000320619.6 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr8_-_93029865 | 0.03 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_-_33686743 | 0.03 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr17_-_5321549 | 0.03 |
ENST00000572809.1 |
NUP88 |
nucleoporin 88kDa |
| chr3_+_35722424 | 0.03 |
ENST00000396481.2 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr12_+_49297899 | 0.03 |
ENST00000552942.1 ENST00000320516.4 |
CCDC65 |
coiled-coil domain containing 65 |
| chr13_-_70682590 | 0.03 |
ENST00000377844.4 |
KLHL1 |
kelch-like family member 1 |
| chr3_+_125694347 | 0.03 |
ENST00000505382.1 ENST00000511082.1 |
ROPN1B |
rhophilin associated tail protein 1B |
| chr4_-_110723194 | 0.03 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr12_-_51718436 | 0.03 |
ENST00000544402.1 |
BIN2 |
bridging integrator 2 |
| chr22_+_30476163 | 0.03 |
ENST00000336726.6 |
HORMAD2 |
HORMA domain containing 2 |
| chrY_-_20935572 | 0.03 |
ENST00000382852.1 ENST00000344884.4 ENST00000304790.3 |
HSFY2 |
heat shock transcription factor, Y linked 2 |
| chr12_-_42631529 | 0.03 |
ENST00000548917.1 |
YAF2 |
YY1 associated factor 2 |
| chr21_-_37914898 | 0.03 |
ENST00000399136.1 |
CLDN14 |
claudin 14 |
| chrY_+_20708557 | 0.03 |
ENST00000307393.2 ENST00000309834.4 ENST00000382856.2 |
HSFY1 |
heat shock transcription factor, Y-linked 1 |
| chr10_-_48806939 | 0.03 |
ENST00000374233.3 ENST00000507417.1 ENST00000512321.1 ENST00000395660.2 ENST00000374235.2 ENST00000395661.3 |
PTPN20B |
protein tyrosine phosphatase, non-receptor type 20B |
| chr12_+_25205568 | 0.03 |
ENST00000548766.1 ENST00000556887.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr10_+_114710211 | 0.03 |
ENST00000349937.2 ENST00000369397.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_-_220264703 | 0.03 |
ENST00000519905.1 ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP |
aspartyl aminopeptidase |
| chr2_+_168043793 | 0.02 |
ENST00000409273.1 ENST00000409605.1 |
XIRP2 |
xin actin-binding repeat containing 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.3 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.5 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.2 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.3 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.0 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.2 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.2 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.3 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 1.0 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.1 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.0 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.0 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.1 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.0 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.9 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |