Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
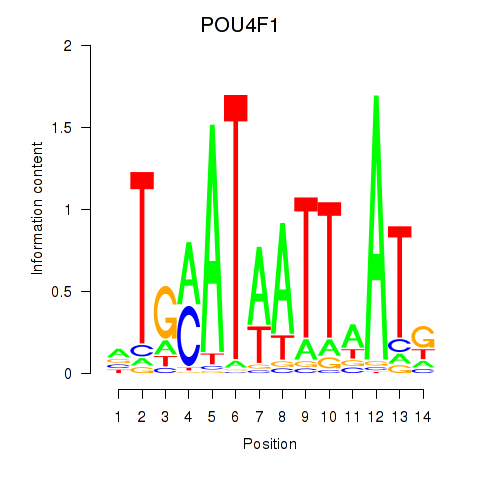
Results for POU4F1_POU4F3
Z-value: 0.38
Transcription factors associated with POU4F1_POU4F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU4F1
|
ENSG00000152192.6 | POU4F1 |
|
POU4F3
|
ENSG00000091010.4 | POU4F3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU4F3 | hg19_v2_chr5_+_145718587_145718607 | -0.69 | 5.8e-02 | Click! |
| POU4F1 | hg19_v2_chr13_-_79177673_79177701 | -0.06 | 8.8e-01 | Click! |
Activity profile of POU4F1_POU4F3 motif
Sorted Z-values of POU4F1_POU4F3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU4F1_POU4F3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_29027696 | 0.64 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr6_-_136847099 | 0.53 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr6_-_136847610 | 0.52 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr17_-_56492989 | 0.35 |
ENST00000583753.1 |
RNF43 |
ring finger protein 43 |
| chrX_-_32173579 | 0.27 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr18_+_47088401 | 0.23 |
ENST00000261292.4 ENST00000427224.2 ENST00000580036.1 |
LIPG |
lipase, endothelial |
| chr19_+_39138320 | 0.18 |
ENST00000424234.2 ENST00000390009.3 ENST00000589528.1 |
ACTN4 |
actinin, alpha 4 |
| chr18_-_52989217 | 0.18 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr4_-_72649763 | 0.18 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr4_-_22444733 | 0.18 |
ENST00000508133.1 |
GPR125 |
G protein-coupled receptor 125 |
| chr12_+_41221794 | 0.17 |
ENST00000547849.1 |
CNTN1 |
contactin 1 |
| chr19_+_39138271 | 0.17 |
ENST00000252699.2 |
ACTN4 |
actinin, alpha 4 |
| chr8_+_98900132 | 0.16 |
ENST00000520016.1 |
MATN2 |
matrilin 2 |
| chr7_-_20256965 | 0.15 |
ENST00000400331.5 ENST00000332878.4 |
MACC1 |
metastasis associated in colon cancer 1 |
| chr1_-_54303949 | 0.14 |
ENST00000234725.8 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr7_-_22234381 | 0.13 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_+_182850551 | 0.13 |
ENST00000452904.1 ENST00000409137.3 ENST00000280295.3 |
PPP1R1C |
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_+_182850743 | 0.13 |
ENST00000409702.1 |
PPP1R1C |
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr19_-_36304201 | 0.13 |
ENST00000301175.3 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr1_+_81771806 | 0.13 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr9_+_35673853 | 0.13 |
ENST00000378357.4 |
CA9 |
carbonic anhydrase IX |
| chr6_+_130339710 | 0.13 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr3_+_98699880 | 0.12 |
ENST00000473756.1 |
LINC00973 |
long intergenic non-protein coding RNA 973 |
| chr1_-_54303934 | 0.12 |
ENST00000537333.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr17_-_10741762 | 0.12 |
ENST00000580256.2 |
PIRT |
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr4_+_154622652 | 0.12 |
ENST00000260010.6 |
TLR2 |
toll-like receptor 2 |
| chr16_-_85784557 | 0.11 |
ENST00000602675.1 |
C16orf74 |
chromosome 16 open reading frame 74 |
| chr1_-_216978709 | 0.11 |
ENST00000360012.3 |
ESRRG |
estrogen-related receptor gamma |
| chr6_-_26108355 | 0.11 |
ENST00000338379.4 |
HIST1H1T |
histone cluster 1, H1t |
| chr9_-_34458531 | 0.10 |
ENST00000379089.1 ENST00000379087.1 ENST00000379084.1 ENST00000379081.1 ENST00000379080.1 ENST00000422409.1 ENST00000379078.1 ENST00000445726.1 ENST00000297620.4 |
FAM219A |
family with sequence similarity 219, member A |
| chr3_+_130569429 | 0.10 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr18_+_28956740 | 0.10 |
ENST00000308128.4 ENST00000359747.4 |
DSG4 |
desmoglein 4 |
| chr3_-_33686743 | 0.10 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr8_-_125577940 | 0.10 |
ENST00000519168.1 ENST00000395508.2 |
MTSS1 |
metastasis suppressor 1 |
| chr3_-_33686925 | 0.10 |
ENST00000485378.2 ENST00000313350.6 ENST00000487200.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr15_-_52587945 | 0.10 |
ENST00000443683.2 ENST00000558479.1 ENST00000261839.7 |
MYO5C |
myosin VC |
| chr12_+_41831485 | 0.10 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr3_-_74570291 | 0.10 |
ENST00000263665.6 |
CNTN3 |
contactin 3 (plasmacytoma associated) |
| chr7_+_150725510 | 0.09 |
ENST00000461373.1 ENST00000358849.4 ENST00000297504.6 ENST00000542328.1 ENST00000498578.1 ENST00000356058.4 ENST00000477719.1 ENST00000477092.1 |
ABCB8 |
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr19_+_17865011 | 0.09 |
ENST00000596462.1 ENST00000596865.1 ENST00000598960.1 ENST00000539407.1 |
FCHO1 |
FCH domain only 1 |
| chr2_-_105882585 | 0.09 |
ENST00000595531.1 |
AC012360.2 |
LOC644617 protein; Uncharacterized protein |
| chr2_-_55237484 | 0.09 |
ENST00000394609.2 |
RTN4 |
reticulon 4 |
| chr6_-_10115007 | 0.08 |
ENST00000485268.1 |
OFCC1 |
orofacial cleft 1 candidate 1 |
| chr13_-_103719196 | 0.08 |
ENST00000245312.3 |
SLC10A2 |
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr15_-_42264702 | 0.08 |
ENST00000220325.4 |
EHD4 |
EH-domain containing 4 |
| chr5_-_77844974 | 0.08 |
ENST00000515007.2 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
| chr1_+_66458072 | 0.08 |
ENST00000423207.2 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr22_+_31488433 | 0.07 |
ENST00000455608.1 |
SMTN |
smoothelin |
| chrX_+_47053208 | 0.07 |
ENST00000442035.1 ENST00000457753.1 ENST00000335972.6 |
UBA1 |
ubiquitin-like modifier activating enzyme 1 |
| chr19_-_55677999 | 0.07 |
ENST00000532817.1 ENST00000527223.2 ENST00000391720.4 |
DNAAF3 |
dynein, axonemal, assembly factor 3 |
| chrX_+_65382433 | 0.07 |
ENST00000374727.3 |
HEPH |
hephaestin |
| chr1_+_161691353 | 0.07 |
ENST00000367948.2 |
FCRLB |
Fc receptor-like B |
| chr14_-_54908043 | 0.07 |
ENST00000556113.1 ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
| chr10_+_695888 | 0.07 |
ENST00000441152.2 |
PRR26 |
proline rich 26 |
| chr19_-_55677920 | 0.07 |
ENST00000524407.2 ENST00000526003.1 ENST00000534170.1 |
DNAAF3 |
dynein, axonemal, assembly factor 3 |
| chr17_-_38821373 | 0.07 |
ENST00000394052.3 |
KRT222 |
keratin 222 |
| chr8_-_62559366 | 0.07 |
ENST00000522919.1 |
ASPH |
aspartate beta-hydroxylase |
| chr2_+_29320571 | 0.06 |
ENST00000401605.1 ENST00000401617.2 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
| chr11_+_35211429 | 0.06 |
ENST00000525688.1 ENST00000278385.6 ENST00000533222.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr2_-_26781521 | 0.06 |
ENST00000403946.3 ENST00000272371.2 |
OTOF |
otoferlin |
| chr10_-_128359074 | 0.06 |
ENST00000544758.1 |
C10orf90 |
chromosome 10 open reading frame 90 |
| chr13_-_46756351 | 0.06 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr12_-_102224704 | 0.06 |
ENST00000299314.7 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr4_+_159236462 | 0.06 |
ENST00000460056.2 |
RXFP1 |
relaxin/insulin-like family peptide receptor 1 |
| chr12_-_102224457 | 0.06 |
ENST00000549165.1 ENST00000549940.1 ENST00000392919.4 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr12_-_89746173 | 0.06 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr1_-_33430286 | 0.06 |
ENST00000373456.7 ENST00000356990.5 ENST00000235150.4 |
RNF19B |
ring finger protein 19B |
| chr3_-_167371740 | 0.06 |
ENST00000466760.1 ENST00000479765.1 |
WDR49 |
WD repeat domain 49 |
| chr12_+_130554803 | 0.05 |
ENST00000535487.1 |
RP11-474D1.2 |
RP11-474D1.2 |
| chr2_-_101034070 | 0.05 |
ENST00000264249.3 |
CHST10 |
carbohydrate sulfotransferase 10 |
| chr17_+_37783453 | 0.05 |
ENST00000579000.1 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr1_-_95538492 | 0.05 |
ENST00000370205.5 |
ALG14 |
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr17_-_38956205 | 0.05 |
ENST00000306658.7 |
KRT28 |
keratin 28 |
| chr3_+_35722487 | 0.05 |
ENST00000441454.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr14_-_65409502 | 0.05 |
ENST00000389614.5 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr10_+_696000 | 0.05 |
ENST00000381489.5 |
PRR26 |
proline rich 26 |
| chr10_-_11574274 | 0.05 |
ENST00000277575.5 |
USP6NL |
USP6 N-terminal like |
| chr7_+_100026406 | 0.05 |
ENST00000414441.1 |
MEPCE |
methylphosphate capping enzyme |
| chr17_+_37783170 | 0.05 |
ENST00000254079.4 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chrX_-_72097698 | 0.05 |
ENST00000373530.1 |
DMRTC1 |
DMRT-like family C1 |
| chr11_+_44117099 | 0.05 |
ENST00000533608.1 |
EXT2 |
exostosin glycosyltransferase 2 |
| chr12_+_54366894 | 0.04 |
ENST00000546378.1 ENST00000243082.4 |
HOXC11 |
homeobox C11 |
| chr19_+_48949030 | 0.04 |
ENST00000253237.5 |
GRWD1 |
glutamate-rich WD repeat containing 1 |
| chr4_+_47487285 | 0.04 |
ENST00000273859.3 ENST00000504445.1 |
ATP10D |
ATPase, class V, type 10D |
| chr1_+_67773044 | 0.04 |
ENST00000262345.1 ENST00000371000.1 |
IL12RB2 |
interleukin 12 receptor, beta 2 |
| chr7_+_99195677 | 0.04 |
ENST00000431679.1 |
GS1-259H13.2 |
GS1-259H13.2 |
| chr11_+_44117260 | 0.04 |
ENST00000358681.4 |
EXT2 |
exostosin glycosyltransferase 2 |
| chr16_-_70835034 | 0.04 |
ENST00000261776.5 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
| chrX_+_72062802 | 0.04 |
ENST00000373533.1 |
DMRTC1B |
DMRT-like family C1B |
| chr18_+_61575200 | 0.04 |
ENST00000238508.3 |
SERPINB10 |
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chrX_+_23682379 | 0.04 |
ENST00000379349.1 |
PRDX4 |
peroxiredoxin 4 |
| chrX_+_41548220 | 0.04 |
ENST00000378142.4 |
GPR34 |
G protein-coupled receptor 34 |
| chrX_+_41548259 | 0.04 |
ENST00000378138.5 |
GPR34 |
G protein-coupled receptor 34 |
| chr2_-_88285309 | 0.04 |
ENST00000420840.2 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
| chrX_-_106146547 | 0.04 |
ENST00000276173.4 ENST00000411805.1 |
RIPPLY1 |
ripply transcriptional repressor 1 |
| chr20_+_30063067 | 0.04 |
ENST00000201979.2 |
REM1 |
RAS (RAD and GEM)-like GTP-binding 1 |
| chr1_+_73771844 | 0.04 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr3_+_70048881 | 0.04 |
ENST00000483525.1 |
RP11-460N16.1 |
RP11-460N16.1 |
| chr6_-_127840336 | 0.04 |
ENST00000525778.1 |
SOGA3 |
SOGA family member 3 |
| chr9_-_74675521 | 0.04 |
ENST00000377024.3 |
C9orf57 |
chromosome 9 open reading frame 57 |
| chr6_-_127840021 | 0.04 |
ENST00000465909.2 |
SOGA3 |
SOGA family member 3 |
| chr2_+_65663812 | 0.04 |
ENST00000606978.1 ENST00000377977.3 ENST00000536804.1 |
AC074391.1 |
AC074391.1 |
| chrX_+_78003204 | 0.03 |
ENST00000435339.3 ENST00000514744.1 |
LPAR4 |
lysophosphatidic acid receptor 4 |
| chr17_+_28268623 | 0.03 |
ENST00000394835.3 ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5 |
EF-hand calcium binding domain 5 |
| chr5_+_139739772 | 0.03 |
ENST00000506757.2 ENST00000230993.6 ENST00000506545.1 ENST00000432095.2 ENST00000507527.1 |
SLC4A9 |
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr2_+_197577841 | 0.03 |
ENST00000409270.1 |
CCDC150 |
coiled-coil domain containing 150 |
| chr7_-_100425112 | 0.03 |
ENST00000358173.3 |
EPHB4 |
EPH receptor B4 |
| chr20_+_36405665 | 0.03 |
ENST00000373469.1 |
CTNNBL1 |
catenin, beta like 1 |
| chrX_-_15288154 | 0.03 |
ENST00000380483.3 ENST00000380485.3 ENST00000380488.4 |
ASB9 |
ankyrin repeat and SOCS box containing 9 |
| chr4_+_106631966 | 0.03 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr2_+_133174147 | 0.03 |
ENST00000329321.3 |
GPR39 |
G protein-coupled receptor 39 |
| chr22_-_38506619 | 0.03 |
ENST00000332536.5 ENST00000381669.3 |
BAIAP2L2 |
BAI1-associated protein 2-like 2 |
| chr3_-_149375783 | 0.03 |
ENST00000467467.1 ENST00000460517.1 ENST00000360632.3 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr4_-_103998439 | 0.03 |
ENST00000503230.1 ENST00000503818.1 |
SLC9B2 |
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr16_+_619931 | 0.03 |
ENST00000321878.5 ENST00000439574.1 ENST00000026218.5 ENST00000470411.2 |
PIGQ |
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr4_+_74269956 | 0.03 |
ENST00000295897.4 ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB |
albumin |
| chr2_+_29336855 | 0.03 |
ENST00000404424.1 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
| chr14_+_22977587 | 0.03 |
ENST00000390504.1 |
TRAJ33 |
T cell receptor alpha joining 33 |
| chr14_+_39703112 | 0.03 |
ENST00000555143.1 ENST00000280082.3 |
MIA2 |
melanoma inhibitory activity 2 |
| chr4_-_122744998 | 0.03 |
ENST00000274026.5 |
CCNA2 |
cyclin A2 |
| chr1_-_197036364 | 0.03 |
ENST00000367412.1 |
F13B |
coagulation factor XIII, B polypeptide |
| chr12_+_100750846 | 0.03 |
ENST00000323346.5 |
SLC17A8 |
solute carrier family 17 (vesicular glutamate transporter), member 8 |
| chr13_-_20110902 | 0.03 |
ENST00000390680.2 ENST00000382977.4 ENST00000382975.4 ENST00000457266.2 |
TPTE2 |
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr6_-_44281043 | 0.03 |
ENST00000244571.4 |
AARS2 |
alanyl-tRNA synthetase 2, mitochondrial |
| chr3_-_194072019 | 0.03 |
ENST00000429275.1 ENST00000323830.3 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
| chr5_+_61874562 | 0.03 |
ENST00000334994.5 ENST00000409534.1 |
LRRC70 IPO11 |
leucine rich repeat containing 70 importin 11 |
| chr3_+_160939050 | 0.03 |
ENST00000493066.1 ENST00000351193.2 ENST00000472947.1 ENST00000463518.1 |
NMD3 |
NMD3 ribosome export adaptor |
| chr1_+_63063152 | 0.03 |
ENST00000371129.3 |
ANGPTL3 |
angiopoietin-like 3 |
| chr10_-_126716459 | 0.03 |
ENST00000309035.6 |
CTBP2 |
C-terminal binding protein 2 |
| chr14_-_21270995 | 0.02 |
ENST00000555698.1 ENST00000397970.4 ENST00000340900.3 |
RNASE1 |
ribonuclease, RNase A family, 1 (pancreatic) |
| chr18_-_33647487 | 0.02 |
ENST00000590898.1 ENST00000357384.4 ENST00000319040.6 ENST00000588737.1 ENST00000399022.4 |
RPRD1A |
regulation of nuclear pre-mRNA domain containing 1A |
| chr15_+_76352178 | 0.02 |
ENST00000388942.3 |
C15orf27 |
chromosome 15 open reading frame 27 |
| chr4_-_68749699 | 0.02 |
ENST00000545541.1 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr7_-_5465045 | 0.02 |
ENST00000399434.2 |
TNRC18 |
trinucleotide repeat containing 18 |
| chr8_+_132916318 | 0.02 |
ENST00000254624.5 ENST00000522709.1 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
| chr1_+_165864800 | 0.02 |
ENST00000469256.2 |
UCK2 |
uridine-cytidine kinase 2 |
| chr7_-_100026280 | 0.02 |
ENST00000360951.4 ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1 |
zinc finger, CW type with PWWP domain 1 |
| chr3_+_35722424 | 0.02 |
ENST00000396481.2 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr4_+_30723003 | 0.02 |
ENST00000543491.1 |
PCDH7 |
protocadherin 7 |
| chr19_+_16940198 | 0.02 |
ENST00000248054.5 ENST00000596802.1 ENST00000379803.1 |
SIN3B |
SIN3 transcription regulator family member B |
| chr5_-_11588907 | 0.02 |
ENST00000513598.1 ENST00000503622.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr8_+_11666649 | 0.02 |
ENST00000528643.1 ENST00000525777.1 |
FDFT1 |
farnesyl-diphosphate farnesyltransferase 1 |
| chr14_+_22580233 | 0.02 |
ENST00000390454.2 |
TRAV25 |
T cell receptor alpha variable 25 |
| chr5_-_11589131 | 0.02 |
ENST00000511377.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr14_+_22675388 | 0.02 |
ENST00000390461.2 |
TRAV34 |
T cell receptor alpha variable 34 |
| chr17_-_5321549 | 0.02 |
ENST00000572809.1 |
NUP88 |
nucleoporin 88kDa |
| chr14_-_21270561 | 0.02 |
ENST00000412779.2 |
RNASE1 |
ribonuclease, RNase A family, 1 (pancreatic) |
| chr17_+_45728427 | 0.02 |
ENST00000540627.1 |
KPNB1 |
karyopherin (importin) beta 1 |
| chr1_+_165864821 | 0.02 |
ENST00000470820.1 |
UCK2 |
uridine-cytidine kinase 2 |
| chr3_-_143567262 | 0.02 |
ENST00000474151.1 ENST00000316549.6 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr11_+_74459876 | 0.02 |
ENST00000299563.4 |
RNF169 |
ring finger protein 169 |
| chr3_+_46742823 | 0.02 |
ENST00000326431.3 |
TMIE |
transmembrane inner ear |
| chr2_-_209054709 | 0.02 |
ENST00000449053.1 ENST00000451346.1 ENST00000341287.4 |
C2orf80 |
chromosome 2 open reading frame 80 |
| chr6_-_161695074 | 0.02 |
ENST00000457520.2 ENST00000366906.5 ENST00000320285.4 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr12_-_11139511 | 0.02 |
ENST00000506868.1 |
TAS2R50 |
taste receptor, type 2, member 50 |
| chr5_+_129083772 | 0.02 |
ENST00000564719.1 |
KIAA1024L |
KIAA1024-like |
| chr11_-_83393457 | 0.02 |
ENST00000404783.3 |
DLG2 |
discs, large homolog 2 (Drosophila) |
| chr14_+_39703084 | 0.02 |
ENST00000553728.1 |
RP11-407N17.3 |
cTAGE family member 5 isoform 4 |
| chr5_-_59481406 | 0.02 |
ENST00000546160.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr22_+_41777927 | 0.02 |
ENST00000266304.4 |
TEF |
thyrotrophic embryonic factor |
| chr2_+_133874577 | 0.02 |
ENST00000596384.1 |
AC011755.1 |
HCG2006742; Protein LOC100996685 |
| chr3_-_69129501 | 0.02 |
ENST00000540295.1 ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3 |
ubiquitin-like modifier activating enzyme 3 |
| chr9_+_135906076 | 0.02 |
ENST00000372097.5 ENST00000440319.1 |
GTF3C5 |
general transcription factor IIIC, polypeptide 5, 63kDa |
| chr2_+_87135076 | 0.02 |
ENST00000409776.2 |
RGPD1 |
RANBP2-like and GRIP domain containing 1 |
| chr1_-_156470515 | 0.02 |
ENST00000340875.5 ENST00000368240.2 ENST00000353795.3 |
MEF2D |
myocyte enhancer factor 2D |
| chr11_+_5509915 | 0.02 |
ENST00000322641.5 |
OR52D1 |
olfactory receptor, family 52, subfamily D, member 1 |
| chrX_-_107975917 | 0.02 |
ENST00000563887.1 |
RP6-24A23.6 |
Uncharacterized protein |
| chr22_+_40322623 | 0.02 |
ENST00000399090.2 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr18_-_31803169 | 0.02 |
ENST00000590712.1 |
NOL4 |
nucleolar protein 4 |
| chr3_-_196242233 | 0.02 |
ENST00000397537.2 |
SMCO1 |
single-pass membrane protein with coiled-coil domains 1 |
| chr6_-_161695042 | 0.02 |
ENST00000366908.5 ENST00000366911.5 ENST00000366905.3 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr11_+_36317830 | 0.02 |
ENST00000530639.1 |
PRR5L |
proline rich 5 like |
| chr12_+_122880045 | 0.02 |
ENST00000539034.1 ENST00000535976.1 |
RP11-450K4.1 |
RP11-450K4.1 |
| chr22_+_40322595 | 0.02 |
ENST00000420971.1 ENST00000544756.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr12_-_111926342 | 0.02 |
ENST00000389154.3 |
ATXN2 |
ataxin 2 |
| chr22_-_43010928 | 0.02 |
ENST00000348657.2 ENST00000252115.5 |
POLDIP3 |
polymerase (DNA-directed), delta interacting protein 3 |
| chr10_+_33271469 | 0.02 |
ENST00000414157.1 |
RP11-462L8.1 |
RP11-462L8.1 |
| chr10_+_135207598 | 0.02 |
ENST00000477902.2 |
MTG1 |
mitochondrial ribosome-associated GTPase 1 |
| chr14_+_24407940 | 0.01 |
ENST00000354854.1 |
DHRS4-AS1 |
DHRS4-AS1 |
| chr2_+_202047843 | 0.01 |
ENST00000272879.5 ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr12_-_23737534 | 0.01 |
ENST00000396007.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr4_-_38784572 | 0.01 |
ENST00000308973.4 ENST00000361424.2 |
TLR10 |
toll-like receptor 10 |
| chr2_+_169757750 | 0.01 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr17_-_49124230 | 0.01 |
ENST00000510283.1 ENST00000510855.1 |
SPAG9 |
sperm associated antigen 9 |
| chr13_+_46039037 | 0.01 |
ENST00000349995.5 |
COG3 |
component of oligomeric golgi complex 3 |
| chr11_-_107328527 | 0.01 |
ENST00000282251.5 ENST00000433523.1 |
CWF19L2 |
CWF19-like 2, cell cycle control (S. pombe) |
| chr12_-_10978957 | 0.01 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr6_+_143929307 | 0.01 |
ENST00000427704.2 ENST00000305766.6 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr7_-_107880508 | 0.01 |
ENST00000425651.2 |
NRCAM |
neuronal cell adhesion molecule |
| chr5_+_55149150 | 0.01 |
ENST00000297015.3 |
IL31RA |
interleukin 31 receptor A |
| chr14_+_39583427 | 0.01 |
ENST00000308317.6 ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2 |
gem (nuclear organelle) associated protein 2 |
| chr4_-_25032501 | 0.01 |
ENST00000382114.4 |
LGI2 |
leucine-rich repeat LGI family, member 2 |
| chr18_-_31803435 | 0.01 |
ENST00000589544.1 ENST00000269185.4 ENST00000261592.5 |
NOL4 |
nucleolar protein 4 |
| chr17_+_18759612 | 0.01 |
ENST00000432893.2 ENST00000414602.1 ENST00000574522.1 ENST00000570450.1 ENST00000419071.2 |
PRPSAP2 |
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr15_-_65426174 | 0.01 |
ENST00000204549.4 |
PDCD7 |
programmed cell death 7 |
| chr1_+_205197304 | 0.01 |
ENST00000358024.3 |
TMCC2 |
transmembrane and coiled-coil domain family 2 |
| chr1_+_47603109 | 0.01 |
ENST00000371890.3 ENST00000294337.3 ENST00000371891.3 |
CYP4A22 |
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr11_+_86085778 | 0.01 |
ENST00000354755.1 ENST00000278487.3 ENST00000531271.1 ENST00000445632.2 |
CCDC81 |
coiled-coil domain containing 81 |
| chr4_+_68424434 | 0.01 |
ENST00000265404.2 ENST00000396225.1 |
STAP1 |
signal transducing adaptor family member 1 |
| chr9_-_21166659 | 0.01 |
ENST00000380225.1 |
IFNA21 |
interferon, alpha 21 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.3 | GO:0070631 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.0 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.0 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |