Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
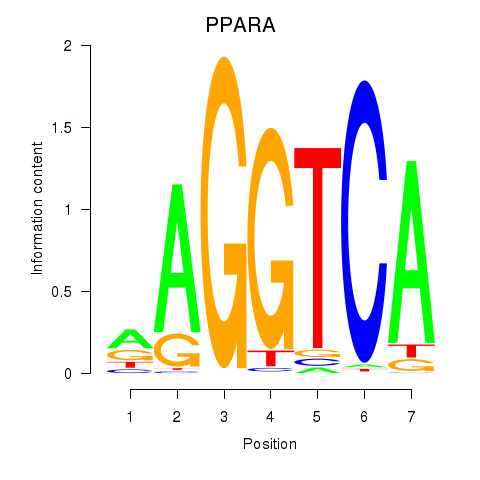
Results for PPARA
Z-value: 0.67
Transcription factors associated with PPARA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARA
|
ENSG00000186951.12 | PPARA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARA | hg19_v2_chr22_+_46546494_46546525 | 0.67 | 7.2e-02 | Click! |
Activity profile of PPARA motif
Sorted Z-values of PPARA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_46640750 | 0.62 |
ENST00000372003.1 |
TSPAN1 |
tetraspanin 1 |
| chr20_+_58179582 | 0.62 |
ENST00000371015.1 ENST00000395639.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr3_+_189349162 | 0.45 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr8_+_86376081 | 0.43 |
ENST00000285379.5 |
CA2 |
carbonic anhydrase II |
| chr9_+_12693336 | 0.41 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr6_+_125524785 | 0.40 |
ENST00000392482.2 |
TPD52L1 |
tumor protein D52-like 1 |
| chr17_+_48609903 | 0.39 |
ENST00000268933.3 |
EPN3 |
epsin 3 |
| chr14_+_76776957 | 0.39 |
ENST00000512784.1 |
ESRRB |
estrogen-related receptor beta |
| chr12_-_53320245 | 0.38 |
ENST00000552150.1 |
KRT8 |
keratin 8 |
| chr15_+_59730348 | 0.34 |
ENST00000288228.5 ENST00000559628.1 ENST00000557914.1 ENST00000560474.1 |
FAM81A |
family with sequence similarity 81, member A |
| chr8_-_144651024 | 0.34 |
ENST00000524906.1 ENST00000532862.1 ENST00000534459.1 |
MROH6 |
maestro heat-like repeat family member 6 |
| chr12_-_12715266 | 0.34 |
ENST00000228862.2 |
DUSP16 |
dual specificity phosphatase 16 |
| chr16_-_68269971 | 0.31 |
ENST00000565858.1 |
ESRP2 |
epithelial splicing regulatory protein 2 |
| chr17_+_48610074 | 0.30 |
ENST00000503690.1 ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3 |
epsin 3 |
| chr2_+_220491973 | 0.30 |
ENST00000358055.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr7_-_44365020 | 0.29 |
ENST00000395747.2 ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chrX_-_31285042 | 0.28 |
ENST00000378680.2 ENST00000378723.3 |
DMD |
dystrophin |
| chr5_+_66124590 | 0.28 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chrX_-_31285018 | 0.28 |
ENST00000361471.4 |
DMD |
dystrophin |
| chrX_-_31284974 | 0.28 |
ENST00000378702.4 |
DMD |
dystrophin |
| chr5_-_60140009 | 0.28 |
ENST00000505959.1 |
ELOVL7 |
ELOVL fatty acid elongase 7 |
| chr2_-_165698662 | 0.28 |
ENST00000194871.6 ENST00000445474.2 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr15_+_45422131 | 0.28 |
ENST00000321429.4 |
DUOX1 |
dual oxidase 1 |
| chr15_+_45422178 | 0.27 |
ENST00000389037.3 ENST00000558322.1 |
DUOX1 |
dual oxidase 1 |
| chr11_-_108408895 | 0.27 |
ENST00000443411.1 ENST00000533052.1 |
EXPH5 |
exophilin 5 |
| chr5_+_145317356 | 0.27 |
ENST00000511217.1 |
SH3RF2 |
SH3 domain containing ring finger 2 |
| chr6_-_136847099 | 0.27 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr2_+_46524537 | 0.27 |
ENST00000263734.3 |
EPAS1 |
endothelial PAS domain protein 1 |
| chr12_-_12674032 | 0.26 |
ENST00000298573.4 |
DUSP16 |
dual specificity phosphatase 16 |
| chr22_+_21128167 | 0.26 |
ENST00000215727.5 |
SERPIND1 |
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr11_-_19223523 | 0.25 |
ENST00000265968.3 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr2_+_18059906 | 0.25 |
ENST00000304101.4 |
KCNS3 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr5_+_145316120 | 0.25 |
ENST00000359120.4 |
SH3RF2 |
SH3 domain containing ring finger 2 |
| chr8_-_127570603 | 0.25 |
ENST00000304916.3 |
FAM84B |
family with sequence similarity 84, member B |
| chr11_-_123756334 | 0.25 |
ENST00000528595.1 ENST00000375026.2 |
TMEM225 |
transmembrane protein 225 |
| chr1_+_212738676 | 0.24 |
ENST00000366981.4 ENST00000366987.2 |
ATF3 |
activating transcription factor 3 |
| chr2_-_165698521 | 0.24 |
ENST00000409184.3 ENST00000392717.2 ENST00000456693.1 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr3_-_141747950 | 0.24 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr15_-_44486632 | 0.23 |
ENST00000484674.1 |
FRMD5 |
FERM domain containing 5 |
| chr19_+_35629702 | 0.22 |
ENST00000351325.4 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr1_-_156786530 | 0.22 |
ENST00000368198.3 |
SH2D2A |
SH2 domain containing 2A |
| chr1_-_156786634 | 0.22 |
ENST00000392306.2 ENST00000368199.3 |
SH2D2A |
SH2 domain containing 2A |
| chr12_-_52828147 | 0.21 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr17_-_29624343 | 0.21 |
ENST00000247271.4 |
OMG |
oligodendrocyte myelin glycoprotein |
| chr16_-_4401258 | 0.21 |
ENST00000577031.1 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr8_+_104831554 | 0.20 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr17_+_9548845 | 0.20 |
ENST00000570475.1 ENST00000285199.7 |
USP43 |
ubiquitin specific peptidase 43 |
| chr6_-_33548006 | 0.20 |
ENST00000374467.3 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr1_-_23886285 | 0.20 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr2_-_74667612 | 0.20 |
ENST00000305557.5 ENST00000233330.6 |
RTKN |
rhotekin |
| chr9_-_117150243 | 0.20 |
ENST00000374088.3 |
AKNA |
AT-hook transcription factor |
| chr17_+_79670386 | 0.19 |
ENST00000333676.3 ENST00000571730.1 ENST00000541223.1 |
MRPL12 SLC25A10 SLC25A10 |
mitochondrial ribosomal protein L12 Mitochondrial dicarboxylate carrier; Uncharacterized protein; cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr6_-_33547975 | 0.19 |
ENST00000442998.2 ENST00000360661.5 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr8_+_86121448 | 0.19 |
ENST00000520225.1 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr8_+_99956759 | 0.19 |
ENST00000522510.1 ENST00000457907.2 |
OSR2 |
odd-skipped related transciption factor 2 |
| chr16_+_691792 | 0.18 |
ENST00000307650.4 |
FAM195A |
family with sequence similarity 195, member A |
| chr12_-_54779511 | 0.18 |
ENST00000551109.1 ENST00000546970.1 |
ZNF385A |
zinc finger protein 385A |
| chr10_+_81107271 | 0.18 |
ENST00000448165.1 |
PPIF |
peptidylprolyl isomerase F |
| chr10_-_76995675 | 0.18 |
ENST00000469299.1 |
COMTD1 |
catechol-O-methyltransferase domain containing 1 |
| chr19_+_39616410 | 0.18 |
ENST00000602004.1 ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4 |
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr16_-_4466565 | 0.18 |
ENST00000572467.1 ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16 CORO7 |
CORO7-PAM16 readthrough coronin 7 |
| chr11_+_10472223 | 0.18 |
ENST00000396554.3 ENST00000524866.1 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr16_-_4588469 | 0.18 |
ENST00000588381.1 ENST00000563332.2 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr10_-_76995769 | 0.18 |
ENST00000372538.3 |
COMTD1 |
catechol-O-methyltransferase domain containing 1 |
| chr16_-_4401284 | 0.18 |
ENST00000318059.3 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr2_+_230787201 | 0.17 |
ENST00000283946.3 |
FBXO36 |
F-box protein 36 |
| chr2_+_220492287 | 0.17 |
ENST00000273063.6 ENST00000373762.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr18_-_24128496 | 0.17 |
ENST00000417602.1 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
| chr2_+_230787213 | 0.17 |
ENST00000409992.1 |
FBXO36 |
F-box protein 36 |
| chr1_+_33231268 | 0.17 |
ENST00000373480.1 |
KIAA1522 |
KIAA1522 |
| chr22_-_29784519 | 0.17 |
ENST00000357586.2 ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1 |
adaptor-related protein complex 1, beta 1 subunit |
| chr15_-_81616446 | 0.17 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr12_+_93963590 | 0.16 |
ENST00000340600.2 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr7_-_151433342 | 0.16 |
ENST00000433631.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chrX_-_133792480 | 0.16 |
ENST00000359237.4 |
PLAC1 |
placenta-specific 1 |
| chr2_-_235405168 | 0.16 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr4_-_122872909 | 0.16 |
ENST00000379645.3 |
TRPC3 |
transient receptor potential cation channel, subfamily C, member 3 |
| chr7_-_151433393 | 0.16 |
ENST00000492843.1 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr19_+_15052301 | 0.16 |
ENST00000248072.3 |
OR7C2 |
olfactory receptor, family 7, subfamily C, member 2 |
| chr19_+_6464502 | 0.16 |
ENST00000308243.7 |
CRB3 |
crumbs homolog 3 (Drosophila) |
| chrX_+_69672136 | 0.16 |
ENST00000374355.3 |
DLG3 |
discs, large homolog 3 (Drosophila) |
| chr22_+_20105012 | 0.16 |
ENST00000331821.3 ENST00000411892.1 |
RANBP1 |
RAN binding protein 1 |
| chr11_+_10471836 | 0.16 |
ENST00000444303.2 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr11_-_66139199 | 0.15 |
ENST00000357440.2 |
SLC29A2 |
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr12_+_32655110 | 0.15 |
ENST00000546442.1 ENST00000583694.1 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr6_+_53659746 | 0.15 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr14_+_42076765 | 0.15 |
ENST00000298119.4 |
LRFN5 |
leucine rich repeat and fibronectin type III domain containing 5 |
| chr20_-_42816206 | 0.15 |
ENST00000372980.3 |
JPH2 |
junctophilin 2 |
| chr22_-_20255212 | 0.15 |
ENST00000416372.1 |
RTN4R |
reticulon 4 receptor |
| chr20_+_1875110 | 0.15 |
ENST00000400068.3 |
SIRPA |
signal-regulatory protein alpha |
| chr19_-_49140609 | 0.15 |
ENST00000601104.1 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
| chr14_+_61654271 | 0.15 |
ENST00000555185.1 ENST00000557294.1 ENST00000556778.1 |
PRKCH |
protein kinase C, eta |
| chr16_+_57653989 | 0.15 |
ENST00000567835.1 ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr17_+_13972807 | 0.15 |
ENST00000429152.2 ENST00000261643.3 ENST00000536205.1 ENST00000537334.1 |
COX10 |
cytochrome c oxidase assembly homolog 10 (yeast) |
| chr3_-_190167571 | 0.15 |
ENST00000354905.2 |
TMEM207 |
transmembrane protein 207 |
| chr19_-_11689752 | 0.15 |
ENST00000592659.1 ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr16_+_57653854 | 0.15 |
ENST00000568908.1 ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr15_+_36887069 | 0.14 |
ENST00000566807.1 ENST00000567389.1 ENST00000562877.1 |
C15orf41 |
chromosome 15 open reading frame 41 |
| chr1_-_205904950 | 0.14 |
ENST00000340781.4 |
SLC26A9 |
solute carrier family 26 (anion exchanger), member 9 |
| chr11_-_66496430 | 0.14 |
ENST00000533211.1 |
SPTBN2 |
spectrin, beta, non-erythrocytic 2 |
| chr3_-_38691119 | 0.14 |
ENST00000333535.4 ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A |
sodium channel, voltage-gated, type V, alpha subunit |
| chr20_+_1875378 | 0.14 |
ENST00000356025.3 |
SIRPA |
signal-regulatory protein alpha |
| chr2_+_220492116 | 0.14 |
ENST00000373760.2 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr12_-_16759711 | 0.14 |
ENST00000447609.1 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr11_+_57365150 | 0.14 |
ENST00000457869.1 ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1 |
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr14_-_21567009 | 0.14 |
ENST00000556174.1 ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219 |
zinc finger protein 219 |
| chr6_-_34664612 | 0.14 |
ENST00000374023.3 ENST00000374026.3 |
C6orf106 |
chromosome 6 open reading frame 106 |
| chr19_+_41699135 | 0.14 |
ENST00000542619.1 ENST00000600561.1 |
CYP2S1 |
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr10_+_104178946 | 0.14 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr3_+_14444063 | 0.14 |
ENST00000454876.2 ENST00000360861.3 ENST00000416216.2 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr14_-_21491477 | 0.14 |
ENST00000298684.5 ENST00000557169.1 ENST00000553563.1 |
NDRG2 |
NDRG family member 2 |
| chr11_+_1860682 | 0.14 |
ENST00000381906.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr1_+_46769303 | 0.13 |
ENST00000311672.5 |
UQCRH |
ubiquinol-cytochrome c reductase hinge protein |
| chr14_-_21492251 | 0.13 |
ENST00000554398.1 |
NDRG2 |
NDRG family member 2 |
| chr4_+_41614909 | 0.13 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr22_+_20104947 | 0.13 |
ENST00000402752.1 |
RANBP1 |
RAN binding protein 1 |
| chr1_+_152881014 | 0.13 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr19_+_6464243 | 0.13 |
ENST00000600229.1 ENST00000356762.3 |
CRB3 |
crumbs homolog 3 (Drosophila) |
| chr1_-_46769261 | 0.13 |
ENST00000343304.6 |
LRRC41 |
leucine rich repeat containing 41 |
| chr7_+_129015484 | 0.13 |
ENST00000490911.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr15_+_65337708 | 0.13 |
ENST00000334287.2 |
SLC51B |
solute carrier family 51, beta subunit |
| chr2_+_219135115 | 0.13 |
ENST00000248451.3 ENST00000273077.4 |
PNKD |
paroxysmal nonkinesigenic dyskinesia |
| chr2_+_220492373 | 0.13 |
ENST00000317151.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chrX_-_21776281 | 0.13 |
ENST00000379494.3 |
SMPX |
small muscle protein, X-linked |
| chr4_+_146560245 | 0.13 |
ENST00000541599.1 |
MMAA |
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr16_+_777118 | 0.13 |
ENST00000562141.1 |
HAGHL |
hydroxyacylglutathione hydrolase-like |
| chr16_+_776936 | 0.13 |
ENST00000549114.1 ENST00000341413.4 ENST00000562187.1 ENST00000564537.1 |
HAGHL |
hydroxyacylglutathione hydrolase-like |
| chr10_+_102759045 | 0.12 |
ENST00000370220.1 |
LZTS2 |
leucine zipper, putative tumor suppressor 2 |
| chr10_+_81107216 | 0.12 |
ENST00000394579.3 ENST00000225174.3 |
PPIF |
peptidylprolyl isomerase F |
| chr1_+_81771806 | 0.12 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr5_+_175085033 | 0.12 |
ENST00000377291.2 |
HRH2 |
histamine receptor H2 |
| chr4_-_87281196 | 0.12 |
ENST00000359221.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr19_-_33360647 | 0.12 |
ENST00000590341.1 ENST00000587772.1 ENST00000023064.4 |
SLC7A9 |
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr17_-_27503770 | 0.12 |
ENST00000533112.1 |
MYO18A |
myosin XVIIIA |
| chr3_+_57875711 | 0.12 |
ENST00000442599.2 |
SLMAP |
sarcolemma associated protein |
| chr14_-_21492113 | 0.12 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr12_-_6483969 | 0.12 |
ENST00000396966.2 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
| chrX_+_135230712 | 0.12 |
ENST00000535737.1 |
FHL1 |
four and a half LIM domains 1 |
| chr22_+_23248512 | 0.12 |
ENST00000390325.2 |
IGLC3 |
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr9_-_100954910 | 0.12 |
ENST00000375077.4 |
CORO2A |
coronin, actin binding protein, 2A |
| chr6_+_89790459 | 0.12 |
ENST00000369472.1 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr20_-_1373606 | 0.12 |
ENST00000381715.1 ENST00000439640.2 ENST00000381719.3 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr9_+_27109392 | 0.11 |
ENST00000406359.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr12_-_6484376 | 0.11 |
ENST00000360168.3 ENST00000358945.3 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
| chr19_+_54371114 | 0.11 |
ENST00000448420.1 ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM |
myeloid-associated differentiation marker |
| chr12_-_16761007 | 0.11 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr3_+_179322573 | 0.11 |
ENST00000493866.1 ENST00000472629.1 ENST00000482604.1 |
NDUFB5 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr1_+_29213678 | 0.11 |
ENST00000347529.3 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr3_-_113465065 | 0.11 |
ENST00000497255.1 ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr19_-_11639910 | 0.11 |
ENST00000588998.1 ENST00000586149.1 |
ECSIT |
ECSIT signalling integrator |
| chr7_-_140624499 | 0.11 |
ENST00000288602.6 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr3_-_45957088 | 0.11 |
ENST00000539217.1 |
LZTFL1 |
leucine zipper transcription factor-like 1 |
| chrX_-_46187069 | 0.11 |
ENST00000446884.1 |
RP1-30G7.2 |
RP1-30G7.2 |
| chr1_+_154947148 | 0.11 |
ENST00000368436.1 ENST00000308987.5 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr16_+_85646763 | 0.11 |
ENST00000411612.1 ENST00000253458.7 |
GSE1 |
Gse1 coiled-coil protein |
| chr1_+_154947126 | 0.11 |
ENST00000368439.1 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr4_+_24797085 | 0.11 |
ENST00000382120.3 |
SOD3 |
superoxide dismutase 3, extracellular |
| chr12_+_93964158 | 0.11 |
ENST00000549206.1 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr3_-_45957534 | 0.11 |
ENST00000536047.1 |
LZTFL1 |
leucine zipper transcription factor-like 1 |
| chr22_+_20105259 | 0.11 |
ENST00000416427.1 ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1 |
RAN binding protein 1 |
| chr20_+_1875942 | 0.11 |
ENST00000358771.4 |
SIRPA |
signal-regulatory protein alpha |
| chr8_-_131028660 | 0.11 |
ENST00000401979.2 ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr1_+_206858328 | 0.11 |
ENST00000367103.3 |
MAPKAPK2 |
mitogen-activated protein kinase-activated protein kinase 2 |
| chr2_+_103378472 | 0.11 |
ENST00000412401.2 |
TMEM182 |
transmembrane protein 182 |
| chr5_-_149682447 | 0.11 |
ENST00000328668.7 |
ARSI |
arylsulfatase family, member I |
| chr10_-_50970322 | 0.11 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr22_+_18121356 | 0.11 |
ENST00000317582.5 ENST00000543133.1 ENST00000538149.1 ENST00000337612.5 ENST00000493680.1 |
BCL2L13 |
BCL2-like 13 (apoptosis facilitator) |
| chr17_+_73975292 | 0.11 |
ENST00000397640.1 ENST00000416485.1 ENST00000588202.1 ENST00000590676.1 ENST00000586891.1 |
TEN1 |
TEN1 CST complex subunit |
| chr22_+_39101728 | 0.11 |
ENST00000216044.5 ENST00000484657.1 |
GTPBP1 |
GTP binding protein 1 |
| chr6_-_41039567 | 0.10 |
ENST00000468811.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chr19_+_1407733 | 0.10 |
ENST00000592453.1 |
DAZAP1 |
DAZ associated protein 1 |
| chr6_+_139456226 | 0.10 |
ENST00000367658.2 |
HECA |
headcase homolog (Drosophila) |
| chr21_+_43823983 | 0.10 |
ENST00000291535.6 ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A |
ubiquitin associated and SH3 domain containing A |
| chr16_+_6069586 | 0.10 |
ENST00000547372.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr6_+_143999072 | 0.10 |
ENST00000440869.2 ENST00000367582.3 ENST00000451827.2 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr11_+_67798363 | 0.10 |
ENST00000525628.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr15_-_44487408 | 0.10 |
ENST00000402883.1 ENST00000417257.1 |
FRMD5 |
FERM domain containing 5 |
| chr14_+_37126765 | 0.10 |
ENST00000402703.2 |
PAX9 |
paired box 9 |
| chr6_+_33172407 | 0.10 |
ENST00000374662.3 |
HSD17B8 |
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr10_-_50970382 | 0.10 |
ENST00000419399.1 ENST00000432695.1 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr19_-_11639931 | 0.10 |
ENST00000592312.1 ENST00000590480.1 ENST00000585318.1 ENST00000252440.7 ENST00000417981.2 ENST00000270517.7 |
ECSIT |
ECSIT signalling integrator |
| chr8_-_144886321 | 0.10 |
ENST00000526832.1 |
SCRIB |
scribbled planar cell polarity protein |
| chr7_+_129007964 | 0.10 |
ENST00000460109.1 ENST00000474594.1 ENST00000446212.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr1_+_165796753 | 0.10 |
ENST00000367879.4 |
UCK2 |
uridine-cytidine kinase 2 |
| chr6_+_42984723 | 0.10 |
ENST00000332245.8 |
KLHDC3 |
kelch domain containing 3 |
| chr6_+_54172653 | 0.10 |
ENST00000370869.3 |
TINAG |
tubulointerstitial nephritis antigen |
| chr19_-_6433765 | 0.10 |
ENST00000321510.6 |
SLC25A41 |
solute carrier family 25, member 41 |
| chr3_+_184055240 | 0.10 |
ENST00000383847.2 |
FAM131A |
family with sequence similarity 131, member A |
| chr22_+_29469012 | 0.10 |
ENST00000400335.4 ENST00000400338.2 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chr2_+_201936458 | 0.10 |
ENST00000237889.4 |
NDUFB3 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr2_+_74757050 | 0.10 |
ENST00000352222.3 ENST00000437202.1 |
HTRA2 |
HtrA serine peptidase 2 |
| chr4_-_13485937 | 0.10 |
ENST00000330852.5 ENST00000288723.4 ENST00000338176.4 |
RAB28 |
RAB28, member RAS oncogene family |
| chr6_+_12007963 | 0.09 |
ENST00000607445.1 |
RP11-456H18.2 |
RP11-456H18.2 |
| chrX_-_109683446 | 0.09 |
ENST00000372057.1 |
AMMECR1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr17_-_8055747 | 0.09 |
ENST00000317276.4 ENST00000581703.1 |
PER1 |
period circadian clock 1 |
| chr2_-_209028300 | 0.09 |
ENST00000304502.4 |
CRYGA |
crystallin, gamma A |
| chr15_-_65715401 | 0.09 |
ENST00000352385.2 |
IGDCC4 |
immunoglobulin superfamily, DCC subclass, member 4 |
| chr4_-_99850243 | 0.09 |
ENST00000280892.6 ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E |
eukaryotic translation initiation factor 4E |
| chr19_-_2328572 | 0.09 |
ENST00000252622.10 |
LSM7 |
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr17_-_38256973 | 0.09 |
ENST00000246672.3 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.1 | 0.4 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.4 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.5 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.3 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.6 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.2 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.2 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.1 | 0.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.3 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.4 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.3 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.3 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.2 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.4 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.2 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.3 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.2 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.4 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.0 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 | 0.1 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.0 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:2000322 | regulation of glucocorticoid receptor signaling pathway(GO:2000322) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:1900738 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.0 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.3 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.4 | GO:0014733 | regulation of skeletal muscle adaptation(GO:0014733) |
| 0.0 | 0.2 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.2 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.2 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.0 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:1904044 | response to aldosterone(GO:1904044) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.0 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.0 | GO:0072615 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.2 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 0.0 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.0 | 0.0 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.0 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.0 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.0 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.0 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.0 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.0 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.2 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.0 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.1 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.3 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.3 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.0 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.0 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | ST ADRENERGIC | Adrenergic Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |