Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
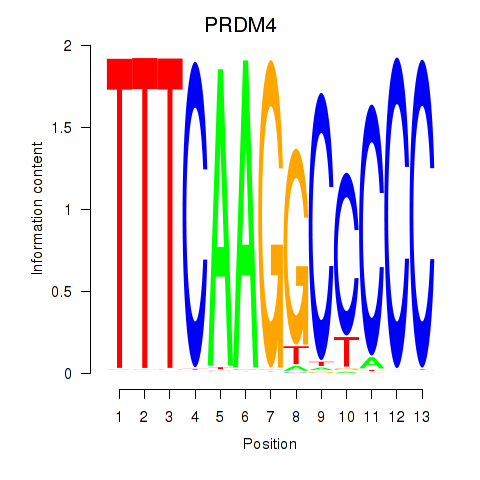
Results for PRDM4
Z-value: 0.32
Transcription factors associated with PRDM4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM4
|
ENSG00000110851.7 | PRDM4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM4 | hg19_v2_chr12_-_108154925_108154945 | -0.82 | 1.2e-02 | Click! |
Activity profile of PRDM4 motif
Sorted Z-values of PRDM4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_62697564 | 0.28 |
ENST00000458442.1 |
TCEA2 |
transcription elongation factor A (SII), 2 |
| chr4_+_667686 | 0.21 |
ENST00000505477.1 |
MYL5 |
myosin, light chain 5, regulatory |
| chr4_+_667307 | 0.20 |
ENST00000506838.1 |
MYL5 |
myosin, light chain 5, regulatory |
| chr22_+_38864041 | 0.18 |
ENST00000216014.4 ENST00000409006.3 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr2_-_179343268 | 0.18 |
ENST00000424785.2 |
FKBP7 |
FK506 binding protein 7 |
| chr14_+_105953204 | 0.17 |
ENST00000409393.2 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
| chr16_+_29823427 | 0.17 |
ENST00000358758.7 ENST00000567659.1 ENST00000572820.1 |
PRRT2 |
proline-rich transmembrane protein 2 |
| chr16_+_29823552 | 0.17 |
ENST00000300797.6 |
PRRT2 |
proline-rich transmembrane protein 2 |
| chr14_+_105953246 | 0.17 |
ENST00000392531.3 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
| chr5_-_61031495 | 0.16 |
ENST00000506550.1 ENST00000512882.2 |
CTD-2170G1.2 |
CTD-2170G1.2 |
| chr2_-_179343226 | 0.16 |
ENST00000434643.2 |
FKBP7 |
FK506 binding protein 7 |
| chrX_-_73072534 | 0.15 |
ENST00000429829.1 |
XIST |
X inactive specific transcript (non-protein coding) |
| chr1_+_43232913 | 0.15 |
ENST00000372525.5 ENST00000536543.1 |
C1orf50 |
chromosome 1 open reading frame 50 |
| chr1_-_43232649 | 0.13 |
ENST00000372526.2 ENST00000236040.4 ENST00000296388.5 ENST00000397054.3 |
LEPRE1 |
leucine proline-enriched proteoglycan (leprecan) 1 |
| chr2_+_219264466 | 0.12 |
ENST00000273062.2 |
CTDSP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr7_-_38398721 | 0.12 |
ENST00000390346.2 |
TRGV3 |
T cell receptor gamma variable 3 |
| chr5_-_180688105 | 0.12 |
ENST00000327767.4 |
TRIM52 |
tripartite motif containing 52 |
| chr19_+_41117770 | 0.12 |
ENST00000601032.1 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr12_+_15475331 | 0.12 |
ENST00000281171.4 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr19_-_49371711 | 0.10 |
ENST00000355496.5 ENST00000263265.6 |
PLEKHA4 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr3_+_16926441 | 0.10 |
ENST00000418129.2 ENST00000396755.2 |
PLCL2 |
phospholipase C-like 2 |
| chr19_-_35264089 | 0.09 |
ENST00000588760.1 ENST00000329285.8 ENST00000587354.2 |
ZNF599 |
zinc finger protein 599 |
| chr9_-_16727978 | 0.09 |
ENST00000418777.1 ENST00000468187.2 |
BNC2 |
basonuclin 2 |
| chr12_+_15475462 | 0.09 |
ENST00000543886.1 ENST00000348962.2 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr7_+_143079000 | 0.08 |
ENST00000392910.2 |
ZYX |
zyxin |
| chr14_+_24025194 | 0.08 |
ENST00000404535.3 ENST00000288014.6 |
THTPA |
thiamine triphosphatase |
| chr11_+_47279504 | 0.07 |
ENST00000441012.2 ENST00000437276.1 ENST00000436029.1 ENST00000467728.1 ENST00000405853.3 |
NR1H3 |
nuclear receptor subfamily 1, group H, member 3 |
| chr6_-_8064567 | 0.07 |
ENST00000543936.1 ENST00000397457.2 |
BLOC1S5 |
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chrX_+_15808569 | 0.07 |
ENST00000380308.3 ENST00000307771.7 |
ZRSR2 |
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
| chr1_+_27114418 | 0.07 |
ENST00000078527.4 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr9_-_70488865 | 0.07 |
ENST00000377392.5 |
CBWD5 |
COBW domain containing 5 |
| chr7_+_143078652 | 0.06 |
ENST00000354434.4 ENST00000449423.2 |
ZYX |
zyxin |
| chr10_-_105212141 | 0.06 |
ENST00000369788.3 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr20_+_30555805 | 0.06 |
ENST00000562532.2 |
XKR7 |
XK, Kell blood group complex subunit-related family, member 7 |
| chr10_+_88728189 | 0.06 |
ENST00000416348.1 |
ADIRF |
adipogenesis regulatory factor |
| chr1_+_27114589 | 0.06 |
ENST00000431541.1 ENST00000449950.2 ENST00000374145.1 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr5_-_78281603 | 0.05 |
ENST00000264914.4 |
ARSB |
arylsulfatase B |
| chr7_+_90893783 | 0.05 |
ENST00000287934.2 |
FZD1 |
frizzled family receptor 1 |
| chr6_-_24721054 | 0.04 |
ENST00000378119.4 |
C6orf62 |
chromosome 6 open reading frame 62 |
| chr8_+_94712732 | 0.04 |
ENST00000518322.1 |
FAM92A1 |
family with sequence similarity 92, member A1 |
| chr12_-_58240470 | 0.04 |
ENST00000548823.1 ENST00000398073.2 |
CTDSP2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr20_-_61569227 | 0.04 |
ENST00000266070.4 ENST00000395335.2 ENST00000266071.5 |
DIDO1 |
death inducer-obliterator 1 |
| chr17_+_42634844 | 0.03 |
ENST00000315323.3 |
FZD2 |
frizzled family receptor 2 |
| chr11_+_58910295 | 0.03 |
ENST00000420244.1 |
FAM111A |
family with sequence similarity 111, member A |
| chr15_+_74287035 | 0.03 |
ENST00000395132.2 ENST00000268059.6 ENST00000354026.6 ENST00000268058.3 ENST00000565898.1 ENST00000569477.1 ENST00000569965.1 ENST00000567543.1 ENST00000436891.3 ENST00000435786.2 ENST00000564428.1 ENST00000359928.4 |
PML |
promyelocytic leukemia |
| chr8_+_94712752 | 0.02 |
ENST00000522324.1 ENST00000522803.1 ENST00000423990.2 |
FAM92A1 |
family with sequence similarity 92, member A1 |
| chr3_+_148709310 | 0.02 |
ENST00000484197.1 ENST00000492285.2 ENST00000461191.1 |
GYG1 |
glycogenin 1 |
| chr3_+_148709128 | 0.02 |
ENST00000345003.4 ENST00000296048.6 ENST00000483267.1 |
GYG1 |
glycogenin 1 |
| chr9_+_17135016 | 0.02 |
ENST00000425824.1 ENST00000262360.5 ENST00000380641.4 |
CNTLN |
centlein, centrosomal protein |
| chr19_-_53636125 | 0.02 |
ENST00000601493.1 ENST00000599261.1 ENST00000597503.1 ENST00000500065.4 ENST00000243643.4 ENST00000594011.1 ENST00000455735.2 ENST00000595193.1 ENST00000448501.1 ENST00000421033.1 ENST00000440291.1 ENST00000595813.1 ENST00000600574.1 ENST00000596051.1 ENST00000601110.1 |
ZNF415 |
zinc finger protein 415 |
| chr16_+_15068955 | 0.02 |
ENST00000396410.4 ENST00000569715.1 ENST00000450288.2 |
PDXDC1 |
pyridoxal-dependent decarboxylase domain containing 1 |
| chr5_+_102455968 | 0.01 |
ENST00000358359.3 |
PPIP5K2 |
diphosphoinositol pentakisphosphate kinase 2 |
| chr2_-_8723918 | 0.01 |
ENST00000454224.1 |
AC011747.4 |
AC011747.4 |
| chr5_-_102455801 | 0.01 |
ENST00000508629.1 ENST00000399004.2 |
GIN1 |
gypsy retrotransposon integrase 1 |
| chr19_+_42724423 | 0.01 |
ENST00000301215.3 ENST00000597945.1 |
ZNF526 |
zinc finger protein 526 |
| chr5_+_102455853 | 0.00 |
ENST00000515845.1 ENST00000321521.9 ENST00000507921.1 |
PPIP5K2 |
diphosphoinositol pentakisphosphate kinase 2 |
| chr10_-_105677886 | 0.00 |
ENST00000224950.3 |
OBFC1 |
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr11_+_8040739 | 0.00 |
ENST00000534099.1 |
TUB |
tubby bipartite transcription factor |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.3 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.3 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.0 | GO:0035425 | autocrine signaling(GO:0035425) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.0 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |