Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
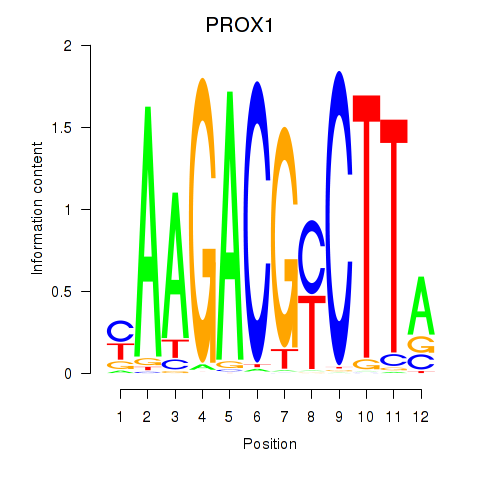
Results for PROX1
Z-value: 0.70
Transcription factors associated with PROX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PROX1
|
ENSG00000117707.11 | PROX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PROX1 | hg19_v2_chr1_+_214163033_214163073 | 0.60 | 1.2e-01 | Click! |
Activity profile of PROX1 motif
Sorted Z-values of PROX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PROX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_82266053 | 0.98 |
ENST00000370715.1 ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2 |
latrophilin 2 |
| chr10_-_123357598 | 0.86 |
ENST00000358487.5 ENST00000369058.3 ENST00000369060.4 ENST00000359354.2 |
FGFR2 |
fibroblast growth factor receptor 2 |
| chr7_-_143105941 | 0.82 |
ENST00000275815.3 |
EPHA1 |
EPH receptor A1 |
| chr9_+_93564191 | 0.74 |
ENST00000375747.1 |
SYK |
spleen tyrosine kinase |
| chr9_+_132096166 | 0.73 |
ENST00000436710.1 |
RP11-65J3.1 |
RP11-65J3.1 |
| chr8_-_41166953 | 0.69 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr22_-_39640756 | 0.61 |
ENST00000331163.6 |
PDGFB |
platelet-derived growth factor beta polypeptide |
| chr11_+_1860200 | 0.49 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr16_+_4845379 | 0.42 |
ENST00000588606.1 ENST00000586005.1 |
SMIM22 |
small integral membrane protein 22 |
| chr6_-_107235287 | 0.38 |
ENST00000436659.1 ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1 |
RP1-60O19.1 |
| chr2_+_163175394 | 0.37 |
ENST00000446271.1 ENST00000429691.2 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr17_+_55183261 | 0.35 |
ENST00000576295.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr11_+_60609537 | 0.35 |
ENST00000227520.5 |
CCDC86 |
coiled-coil domain containing 86 |
| chr7_+_48128316 | 0.34 |
ENST00000341253.4 |
UPP1 |
uridine phosphorylase 1 |
| chr11_+_117947724 | 0.33 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr7_+_48128194 | 0.33 |
ENST00000416681.1 ENST00000331803.4 ENST00000432131.1 |
UPP1 |
uridine phosphorylase 1 |
| chr1_+_70876926 | 0.33 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr1_+_171454659 | 0.32 |
ENST00000367742.3 ENST00000338920.4 |
PRRC2C |
proline-rich coiled-coil 2C |
| chr16_+_50300427 | 0.32 |
ENST00000394697.2 ENST00000566433.2 ENST00000538642.1 |
ADCY7 |
adenylate cyclase 7 |
| chr3_+_136676707 | 0.32 |
ENST00000329582.4 |
IL20RB |
interleukin 20 receptor beta |
| chr1_+_171454639 | 0.31 |
ENST00000392078.3 ENST00000426496.2 |
PRRC2C |
proline-rich coiled-coil 2C |
| chr1_+_70876891 | 0.31 |
ENST00000411986.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr14_+_45605157 | 0.31 |
ENST00000542564.2 |
FANCM |
Fanconi anemia, complementation group M |
| chr11_+_117947782 | 0.30 |
ENST00000522307.1 ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr14_+_23842018 | 0.29 |
ENST00000397242.2 ENST00000329715.2 |
IL25 |
interleukin 25 |
| chr6_+_111580508 | 0.28 |
ENST00000368847.4 |
KIAA1919 |
KIAA1919 |
| chrX_-_153237258 | 0.28 |
ENST00000310441.7 |
HCFC1 |
host cell factor C1 (VP16-accessory protein) |
| chr17_+_41994576 | 0.28 |
ENST00000588043.2 |
FAM215A |
family with sequence similarity 215, member A (non-protein coding) |
| chrX_-_153236819 | 0.26 |
ENST00000354233.3 |
HCFC1 |
host cell factor C1 (VP16-accessory protein) |
| chrX_-_153236620 | 0.26 |
ENST00000369984.4 |
HCFC1 |
host cell factor C1 (VP16-accessory protein) |
| chr4_+_26862431 | 0.26 |
ENST00000465503.1 |
STIM2 |
stromal interaction molecule 2 |
| chr6_-_90529418 | 0.25 |
ENST00000439638.1 ENST00000369393.3 ENST00000428876.1 |
MDN1 |
MDN1, midasin homolog (yeast) |
| chr2_+_97001491 | 0.25 |
ENST00000240423.4 ENST00000427946.1 ENST00000435975.1 ENST00000456906.1 ENST00000455200.1 |
NCAPH |
non-SMC condensin I complex, subunit H |
| chr4_-_170947522 | 0.25 |
ENST00000361618.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr4_+_26862400 | 0.24 |
ENST00000467011.1 ENST00000412829.2 |
STIM2 |
stromal interaction molecule 2 |
| chr8_+_104310661 | 0.24 |
ENST00000522566.1 |
FZD6 |
frizzled family receptor 6 |
| chr22_-_50050986 | 0.24 |
ENST00000405854.1 |
C22orf34 |
chromosome 22 open reading frame 34 |
| chr14_+_78266408 | 0.23 |
ENST00000238561.5 |
ADCK1 |
aarF domain containing kinase 1 |
| chr1_+_179262905 | 0.23 |
ENST00000539888.1 ENST00000540564.1 ENST00000535686.1 ENST00000367619.3 |
SOAT1 |
sterol O-acyltransferase 1 |
| chr16_-_28857677 | 0.22 |
ENST00000313511.3 |
TUFM |
Tu translation elongation factor, mitochondrial |
| chr2_-_89247338 | 0.22 |
ENST00000496168.1 |
IGKV1-5 |
immunoglobulin kappa variable 1-5 |
| chr16_-_10674528 | 0.22 |
ENST00000359543.3 |
EMP2 |
epithelial membrane protein 2 |
| chr18_-_21166841 | 0.22 |
ENST00000269228.5 |
NPC1 |
Niemann-Pick disease, type C1 |
| chr1_-_246729544 | 0.22 |
ENST00000544618.1 ENST00000366514.4 |
TFB2M |
transcription factor B2, mitochondrial |
| chr4_+_26862313 | 0.21 |
ENST00000467087.1 ENST00000382009.3 ENST00000237364.5 |
STIM2 |
stromal interaction molecule 2 |
| chr1_-_204380919 | 0.21 |
ENST00000367188.4 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
| chr12_+_113229543 | 0.21 |
ENST00000447659.2 |
RPH3A |
rabphilin 3A homolog (mouse) |
| chr14_+_45605127 | 0.21 |
ENST00000556036.1 ENST00000267430.5 |
FANCM |
Fanconi anemia, complementation group M |
| chr11_-_59383617 | 0.21 |
ENST00000263847.1 |
OSBP |
oxysterol binding protein |
| chr12_+_113229452 | 0.20 |
ENST00000389385.4 |
RPH3A |
rabphilin 3A homolog (mouse) |
| chr1_-_23342340 | 0.20 |
ENST00000566855.1 |
C1orf234 |
chromosome 1 open reading frame 234 |
| chr18_+_33767473 | 0.20 |
ENST00000261326.5 |
MOCOS |
molybdenum cofactor sulfurase |
| chr6_-_4079334 | 0.20 |
ENST00000492651.1 ENST00000498677.1 ENST00000274673.3 |
FAM217A |
family with sequence similarity 217, member A |
| chr19_+_45754505 | 0.20 |
ENST00000262891.4 ENST00000300843.4 |
MARK4 |
MAP/microtubule affinity-regulating kinase 4 |
| chr12_+_20522179 | 0.19 |
ENST00000359062.3 |
PDE3A |
phosphodiesterase 3A, cGMP-inhibited |
| chr4_-_38666430 | 0.19 |
ENST00000436901.1 |
AC021860.1 |
Uncharacterized protein |
| chr11_-_118023490 | 0.19 |
ENST00000324727.4 |
SCN4B |
sodium channel, voltage-gated, type IV, beta subunit |
| chr10_-_96122682 | 0.19 |
ENST00000371361.3 |
NOC3L |
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr8_+_110552831 | 0.18 |
ENST00000530629.1 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
| chr5_+_145826867 | 0.18 |
ENST00000296702.5 ENST00000394421.2 |
TCERG1 |
transcription elongation regulator 1 |
| chr4_-_170947485 | 0.18 |
ENST00000504999.1 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr8_+_110552046 | 0.17 |
ENST00000529931.1 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
| chr9_-_130497565 | 0.17 |
ENST00000336067.6 ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A |
torsin family 2, member A |
| chr2_-_239140276 | 0.17 |
ENST00000334973.4 |
AC016757.3 |
Protein LOC151174 |
| chr19_+_4007644 | 0.17 |
ENST00000262971.2 |
PIAS4 |
protein inhibitor of activated STAT, 4 |
| chr12_+_113229737 | 0.17 |
ENST00000551052.1 ENST00000415485.3 |
RPH3A |
rabphilin 3A homolog (mouse) |
| chr22_+_26138108 | 0.17 |
ENST00000536101.1 ENST00000335473.7 ENST00000407587.2 |
MYO18B |
myosin XVIIIB |
| chr15_-_31453157 | 0.17 |
ENST00000559177.1 ENST00000558445.1 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
| chr17_+_72209653 | 0.16 |
ENST00000269346.4 |
TTYH2 |
tweety family member 2 |
| chr4_-_170948361 | 0.16 |
ENST00000393702.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr10_+_16478942 | 0.16 |
ENST00000535784.2 ENST00000423462.2 ENST00000378000.1 |
PTER |
phosphotriesterase related |
| chr6_+_158957431 | 0.16 |
ENST00000367090.3 |
TMEM181 |
transmembrane protein 181 |
| chr11_+_6518503 | 0.16 |
ENST00000496802.1 ENST00000254579.6 ENST00000354685.3 |
DNHD1 |
dynein heavy chain domain 1 |
| chr2_-_239140011 | 0.16 |
ENST00000409376.1 ENST00000409070.1 ENST00000409942.1 |
AC016757.3 |
Protein LOC151174 |
| chr17_-_40273348 | 0.16 |
ENST00000225916.5 |
KAT2A |
K(lysine) acetyltransferase 2A |
| chr6_-_31628512 | 0.15 |
ENST00000375911.1 |
C6orf47 |
chromosome 6 open reading frame 47 |
| chr2_-_89310012 | 0.15 |
ENST00000493819.1 |
IGKV1-9 |
immunoglobulin kappa variable 1-9 |
| chr14_+_22782867 | 0.15 |
ENST00000390467.3 |
TRAV40 |
T cell receptor alpha variable 40 |
| chr1_+_179050512 | 0.15 |
ENST00000367627.3 |
TOR3A |
torsin family 3, member A |
| chr6_-_86303833 | 0.14 |
ENST00000505648.1 |
SNX14 |
sorting nexin 14 |
| chr16_+_30669720 | 0.14 |
ENST00000356166.6 |
FBRS |
fibrosin |
| chr11_-_73882029 | 0.14 |
ENST00000539061.1 |
C2CD3 |
C2 calcium-dependent domain containing 3 |
| chr17_-_38074859 | 0.14 |
ENST00000520542.1 ENST00000418519.1 ENST00000394179.1 |
GSDMB |
gasdermin B |
| chr1_-_225616515 | 0.14 |
ENST00000338179.2 ENST00000425080.1 |
LBR |
lamin B receptor |
| chr8_+_145064215 | 0.14 |
ENST00000313269.5 |
GRINA |
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr9_-_86571628 | 0.14 |
ENST00000376344.3 |
C9orf64 |
chromosome 9 open reading frame 64 |
| chr8_+_145064233 | 0.14 |
ENST00000529301.1 ENST00000395068.4 |
GRINA |
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr7_+_150759634 | 0.14 |
ENST00000392826.2 ENST00000461735.1 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
| chr2_+_101869262 | 0.13 |
ENST00000289382.3 |
CNOT11 |
CCR4-NOT transcription complex, subunit 11 |
| chr1_-_94050668 | 0.13 |
ENST00000539242.1 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr10_-_50970322 | 0.13 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr2_-_163175133 | 0.13 |
ENST00000421365.2 ENST00000263642.2 |
IFIH1 |
interferon induced with helicase C domain 1 |
| chr2_-_225434538 | 0.13 |
ENST00000409096.1 |
CUL3 |
cullin 3 |
| chr9_+_125132803 | 0.13 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr11_-_46408107 | 0.13 |
ENST00000433765.2 |
CHRM4 |
cholinergic receptor, muscarinic 4 |
| chr8_-_143696833 | 0.12 |
ENST00000356613.2 |
ARC |
activity-regulated cytoskeleton-associated protein |
| chr8_+_117963190 | 0.12 |
ENST00000427715.2 |
SLC30A8 |
solute carrier family 30 (zinc transporter), member 8 |
| chr5_-_157002775 | 0.12 |
ENST00000257527.4 |
ADAM19 |
ADAM metallopeptidase domain 19 |
| chr6_-_137540477 | 0.12 |
ENST00000367735.2 ENST00000367739.4 ENST00000458076.1 ENST00000414770.1 |
IFNGR1 |
interferon gamma receptor 1 |
| chr12_-_52946923 | 0.12 |
ENST00000267119.5 |
KRT71 |
keratin 71 |
| chr4_-_170947565 | 0.12 |
ENST00000506764.1 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr7_-_91509986 | 0.12 |
ENST00000456229.1 ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF |
mitochondrial transcription termination factor |
| chr17_-_48546232 | 0.12 |
ENST00000258969.4 |
CHAD |
chondroadherin |
| chr15_-_101835110 | 0.12 |
ENST00000560496.1 |
SNRPA1 |
small nuclear ribonucleoprotein polypeptide A' |
| chr19_+_35773242 | 0.12 |
ENST00000222304.3 |
HAMP |
hepcidin antimicrobial peptide |
| chr11_+_73882311 | 0.12 |
ENST00000398427.4 ENST00000544401.1 |
PPME1 |
protein phosphatase methylesterase 1 |
| chr1_+_35544968 | 0.12 |
ENST00000359858.4 ENST00000373330.1 |
ZMYM1 |
zinc finger, MYM-type 1 |
| chr2_+_128403720 | 0.11 |
ENST00000272644.3 |
GPR17 |
G protein-coupled receptor 17 |
| chr2_+_128403439 | 0.11 |
ENST00000544369.1 |
GPR17 |
G protein-coupled receptor 17 |
| chr11_+_73882144 | 0.11 |
ENST00000328257.8 |
PPME1 |
protein phosphatase methylesterase 1 |
| chr3_+_12525931 | 0.11 |
ENST00000446004.1 ENST00000314571.7 ENST00000454502.2 ENST00000383797.5 ENST00000402228.3 ENST00000284995.6 ENST00000444864.1 |
TSEN2 |
TSEN2 tRNA splicing endonuclease subunit |
| chr10_-_50970382 | 0.11 |
ENST00000419399.1 ENST00000432695.1 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr7_+_99775520 | 0.11 |
ENST00000317296.5 ENST00000422690.1 ENST00000439782.1 |
STAG3 |
stromal antigen 3 |
| chr3_-_197025447 | 0.11 |
ENST00000346964.2 ENST00000357674.4 ENST00000314062.3 ENST00000448528.2 ENST00000419553.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr6_-_7911042 | 0.11 |
ENST00000379757.4 |
TXNDC5 |
thioredoxin domain containing 5 (endoplasmic reticulum) |
| chr1_-_111061797 | 0.11 |
ENST00000369771.2 |
KCNA10 |
potassium voltage-gated channel, shaker-related subfamily, member 10 |
| chr19_-_19754354 | 0.10 |
ENST00000587238.1 |
GMIP |
GEM interacting protein |
| chr12_-_43945724 | 0.10 |
ENST00000389420.3 ENST00000553158.1 |
ADAMTS20 |
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr2_+_17935383 | 0.10 |
ENST00000524465.1 ENST00000381254.2 ENST00000532257.1 |
GEN1 |
GEN1 Holliday junction 5' flap endonuclease |
| chr15_-_41047421 | 0.10 |
ENST00000560460.1 ENST00000338376.3 ENST00000560905.1 |
RMDN3 |
regulator of microtubule dynamics 3 |
| chr11_+_64008525 | 0.10 |
ENST00000449942.2 |
FKBP2 |
FK506 binding protein 2, 13kDa |
| chr1_+_165864821 | 0.10 |
ENST00000470820.1 |
UCK2 |
uridine-cytidine kinase 2 |
| chr11_+_93394805 | 0.10 |
ENST00000325212.6 ENST00000411936.1 ENST00000344196.4 |
KIAA1731 |
KIAA1731 |
| chr19_-_45909585 | 0.10 |
ENST00000593226.1 ENST00000418234.2 |
PPP1R13L |
protein phosphatase 1, regulatory subunit 13 like |
| chr4_-_128887069 | 0.10 |
ENST00000541133.1 ENST00000296468.3 ENST00000513559.1 |
MFSD8 |
major facilitator superfamily domain containing 8 |
| chr19_-_19754404 | 0.10 |
ENST00000587205.1 ENST00000445806.2 ENST00000203556.4 |
GMIP |
GEM interacting protein |
| chr1_+_165864800 | 0.09 |
ENST00000469256.2 |
UCK2 |
uridine-cytidine kinase 2 |
| chr1_+_222817629 | 0.09 |
ENST00000340535.7 |
MIA3 |
melanoma inhibitory activity family, member 3 |
| chr10_-_69455873 | 0.09 |
ENST00000433211.2 |
CTNNA3 |
catenin (cadherin-associated protein), alpha 3 |
| chr19_-_46234119 | 0.09 |
ENST00000317683.3 |
FBXO46 |
F-box protein 46 |
| chr18_-_2571437 | 0.09 |
ENST00000574676.1 ENST00000574538.1 ENST00000319888.6 |
METTL4 |
methyltransferase like 4 |
| chr14_+_24563262 | 0.09 |
ENST00000559250.1 ENST00000216780.4 ENST00000560736.1 ENST00000396973.4 ENST00000559837.1 |
PCK2 |
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr6_+_117198400 | 0.09 |
ENST00000332958.2 |
RFX6 |
regulatory factor X, 6 |
| chr8_-_87755878 | 0.09 |
ENST00000320005.5 |
CNGB3 |
cyclic nucleotide gated channel beta 3 |
| chr6_+_117996621 | 0.09 |
ENST00000368494.3 |
NUS1 |
nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae) |
| chr1_-_207226313 | 0.08 |
ENST00000367084.1 |
YOD1 |
YOD1 deubiquitinase |
| chr21_-_40033618 | 0.08 |
ENST00000417133.2 ENST00000398910.1 ENST00000442448.1 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr12_-_122296755 | 0.08 |
ENST00000289004.4 |
HPD |
4-hydroxyphenylpyruvate dioxygenase |
| chr22_-_45559540 | 0.08 |
ENST00000432502.1 |
CTA-217C2.1 |
CTA-217C2.1 |
| chr6_+_28227063 | 0.08 |
ENST00000343684.3 |
NKAPL |
NFKB activating protein-like |
| chr5_-_157002749 | 0.08 |
ENST00000517905.1 ENST00000430702.2 ENST00000394020.1 |
ADAM19 |
ADAM metallopeptidase domain 19 |
| chr6_+_29141311 | 0.08 |
ENST00000377167.2 |
OR2J2 |
olfactory receptor, family 2, subfamily J, member 2 |
| chr10_+_134258649 | 0.08 |
ENST00000392630.3 ENST00000321248.2 |
C10orf91 |
chromosome 10 open reading frame 91 |
| chr22_-_45559642 | 0.08 |
ENST00000426282.2 |
CTA-217C2.1 |
CTA-217C2.1 |
| chr2_+_28718921 | 0.08 |
ENST00000327757.5 ENST00000422425.2 ENST00000404858.1 |
PLB1 |
phospholipase B1 |
| chr1_-_44497118 | 0.08 |
ENST00000537678.1 ENST00000466926.1 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr2_-_152830479 | 0.08 |
ENST00000360283.6 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr2_+_89999259 | 0.08 |
ENST00000558026.1 |
IGKV2D-28 |
immunoglobulin kappa variable 2D-28 |
| chr3_-_112218205 | 0.08 |
ENST00000383680.4 |
BTLA |
B and T lymphocyte associated |
| chr1_-_44497024 | 0.08 |
ENST00000372306.3 ENST00000372310.3 ENST00000475075.2 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr11_+_46740730 | 0.08 |
ENST00000311907.5 ENST00000530231.1 ENST00000442468.1 |
F2 |
coagulation factor II (thrombin) |
| chr18_+_60190226 | 0.08 |
ENST00000269499.5 |
ZCCHC2 |
zinc finger, CCHC domain containing 2 |
| chrY_+_20708557 | 0.07 |
ENST00000307393.2 ENST00000309834.4 ENST00000382856.2 |
HSFY1 |
heat shock transcription factor, Y-linked 1 |
| chr4_-_140477353 | 0.07 |
ENST00000406354.1 ENST00000506866.2 |
SETD7 |
SET domain containing (lysine methyltransferase) 7 |
| chr11_+_86106208 | 0.07 |
ENST00000528728.1 |
CCDC81 |
coiled-coil domain containing 81 |
| chrY_-_20935572 | 0.07 |
ENST00000382852.1 ENST00000344884.4 ENST00000304790.3 |
HSFY2 |
heat shock transcription factor, Y linked 2 |
| chr14_+_73603126 | 0.07 |
ENST00000557356.1 ENST00000556864.1 ENST00000556533.1 ENST00000556951.1 ENST00000557293.1 ENST00000553719.1 ENST00000553599.1 ENST00000556011.1 ENST00000394157.3 ENST00000357710.4 ENST00000324501.5 ENST00000560005.2 ENST00000555254.1 ENST00000261970.3 ENST00000344094.3 ENST00000554131.1 ENST00000557037.1 |
PSEN1 |
presenilin 1 |
| chr7_+_99724317 | 0.07 |
ENST00000398075.2 ENST00000421390.1 |
MBLAC1 |
metallo-beta-lactamase domain containing 1 |
| chr11_+_61159832 | 0.07 |
ENST00000334888.5 ENST00000398979.3 |
TMEM216 |
transmembrane protein 216 |
| chr12_+_72079842 | 0.07 |
ENST00000266673.5 ENST00000550524.1 |
TMEM19 |
transmembrane protein 19 |
| chr5_+_179125368 | 0.07 |
ENST00000502296.1 ENST00000504734.1 ENST00000415618.2 |
CANX |
calnexin |
| chr16_+_12070567 | 0.07 |
ENST00000566228.1 |
SNX29 |
sorting nexin 29 |
| chr15_-_78913521 | 0.07 |
ENST00000326828.5 |
CHRNA3 |
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr7_-_25164868 | 0.06 |
ENST00000409409.1 ENST00000409764.1 ENST00000413447.1 |
CYCS |
cytochrome c, somatic |
| chr4_+_128886424 | 0.06 |
ENST00000398965.1 |
C4orf29 |
chromosome 4 open reading frame 29 |
| chr4_+_164265035 | 0.06 |
ENST00000338566.3 |
NPY5R |
neuropeptide Y receptor Y5 |
| chr3_-_197024965 | 0.06 |
ENST00000392382.2 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr4_-_69215467 | 0.06 |
ENST00000579690.1 |
YTHDC1 |
YTH domain containing 1 |
| chr16_+_31885079 | 0.06 |
ENST00000300870.10 ENST00000394846.3 |
ZNF267 |
zinc finger protein 267 |
| chr22_+_45559722 | 0.06 |
ENST00000347635.4 ENST00000407019.2 ENST00000424634.1 ENST00000417702.1 ENST00000425733.2 ENST00000430547.1 |
NUP50 |
nucleoporin 50kDa |
| chr22_-_43539346 | 0.06 |
ENST00000327555.5 ENST00000290429.6 |
MCAT |
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr2_+_232826211 | 0.06 |
ENST00000325385.7 ENST00000360410.4 |
DIS3L2 |
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
| chr10_+_45869652 | 0.06 |
ENST00000542434.1 ENST00000374391.2 |
ALOX5 |
arachidonate 5-lipoxygenase |
| chr15_-_91537723 | 0.06 |
ENST00000394249.3 ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1 |
protein regulator of cytokinesis 1 |
| chr14_-_31676964 | 0.06 |
ENST00000553700.1 |
HECTD1 |
HECT domain containing E3 ubiquitin protein ligase 1 |
| chr7_-_25164969 | 0.06 |
ENST00000305786.2 |
CYCS |
cytochrome c, somatic |
| chr20_+_30028322 | 0.06 |
ENST00000376309.3 |
DEFB123 |
defensin, beta 123 |
| chr16_-_22012419 | 0.05 |
ENST00000537222.2 ENST00000424898.2 ENST00000286143.6 |
PDZD9 |
PDZ domain containing 9 |
| chr9_-_127533519 | 0.05 |
ENST00000487099.2 ENST00000344523.4 ENST00000373584.3 |
NR6A1 |
nuclear receptor subfamily 6, group A, member 1 |
| chr9_+_71789133 | 0.05 |
ENST00000348208.4 ENST00000265384.7 |
TJP2 |
tight junction protein 2 |
| chr16_+_28722684 | 0.05 |
ENST00000331666.6 ENST00000395587.1 ENST00000569690.1 ENST00000564243.1 |
EIF3C |
eukaryotic translation initiation factor 3, subunit C |
| chr19_+_53073526 | 0.05 |
ENST00000596514.1 ENST00000391785.3 ENST00000301093.2 |
ZNF701 |
zinc finger protein 701 |
| chr1_+_95975672 | 0.05 |
ENST00000440116.2 ENST00000456933.1 |
RP11-286B14.1 |
RP11-286B14.1 |
| chr3_-_196065248 | 0.05 |
ENST00000446879.1 ENST00000273695.3 |
TM4SF19 |
transmembrane 4 L six family member 19 |
| chr7_+_47834880 | 0.05 |
ENST00000258776.4 |
C7orf69 |
chromosome 7 open reading frame 69 |
| chr9_+_72873837 | 0.05 |
ENST00000361138.5 |
SMC5 |
structural maintenance of chromosomes 5 |
| chr15_-_76304731 | 0.05 |
ENST00000394907.3 |
NRG4 |
neuregulin 4 |
| chr3_-_137834436 | 0.05 |
ENST00000327532.2 ENST00000467030.1 |
DZIP1L |
DAZ interacting zinc finger protein 1-like |
| chr10_+_104005272 | 0.05 |
ENST00000369983.3 |
GBF1 |
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr15_-_78913628 | 0.05 |
ENST00000348639.3 |
CHRNA3 |
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr21_-_26979786 | 0.04 |
ENST00000419219.1 ENST00000352957.4 ENST00000307301.7 |
MRPL39 |
mitochondrial ribosomal protein L39 |
| chr6_-_32634425 | 0.04 |
ENST00000399082.3 ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1 |
major histocompatibility complex, class II, DQ beta 1 |
| chr16_+_28722809 | 0.04 |
ENST00000566866.1 |
EIF3C |
eukaryotic translation initiation factor 3, subunit C |
| chr17_-_48546324 | 0.04 |
ENST00000508540.1 |
CHAD |
chondroadherin |
| chr12_-_46766577 | 0.04 |
ENST00000256689.5 |
SLC38A2 |
solute carrier family 38, member 2 |
| chr1_-_202311088 | 0.04 |
ENST00000367274.4 |
UBE2T |
ubiquitin-conjugating enzyme E2T (putative) |
| chr19_-_16582815 | 0.04 |
ENST00000455140.2 ENST00000248070.6 ENST00000594975.1 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
| chr19_+_49535169 | 0.04 |
ENST00000474913.1 ENST00000359342.6 |
CGB2 |
chorionic gonadotropin, beta polypeptide 2 |
| chr11_+_61722629 | 0.04 |
ENST00000526988.1 |
BEST1 |
bestrophin 1 |
| chr9_-_127533582 | 0.03 |
ENST00000416460.2 |
NR6A1 |
nuclear receptor subfamily 6, group A, member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.2 | 0.7 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.2 | 0.8 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.2 | 0.7 | GO:0045399 | response to molecule of fungal origin(GO:0002238) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 0.2 | 0.7 | GO:0006218 | uridine catabolic process(GO:0006218) |
| 0.2 | 0.6 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.2 | 0.6 | GO:1905176 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 0.7 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.3 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 0.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.2 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.2 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.2 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.3 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.2 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.1 | GO:1904479 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.2 | GO:0043983 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.2 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.3 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of keratinocyte apoptotic process(GO:1902174) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.2 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.1 | GO:1900738 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0032383 | dolichol metabolic process(GO:0019348) regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.0 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.7 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.2 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 1.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.2 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.8 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.2 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |