Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for PRRX1_ALX4_PHOX2A
Z-value: 1.67
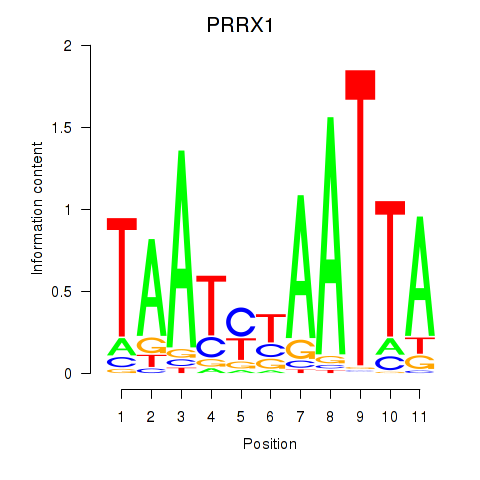
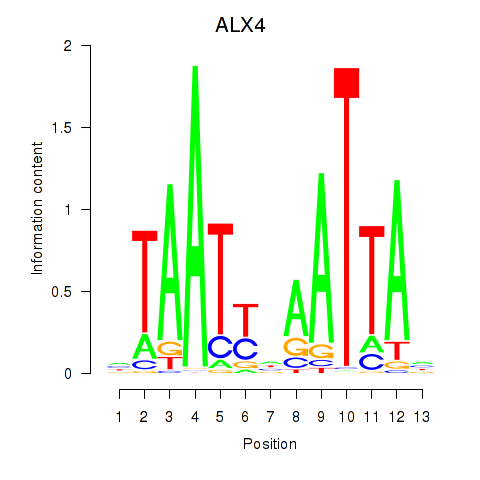
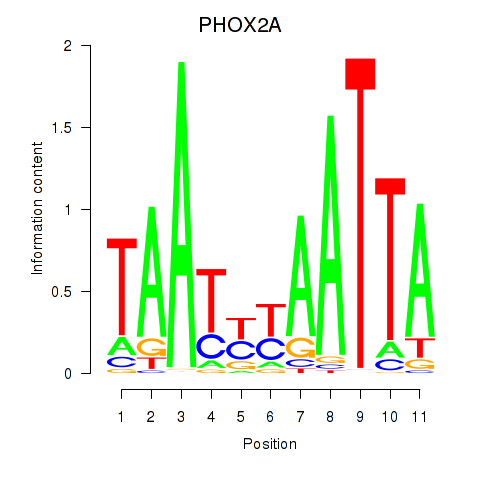
Transcription factors associated with PRRX1_ALX4_PHOX2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRRX1
|
ENSG00000116132.7 | PRRX1 |
|
ALX4
|
ENSG00000052850.5 | ALX4 |
|
PHOX2A
|
ENSG00000165462.5 | PHOX2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRRX1 | hg19_v2_chr1_+_170633047_170633084 | 0.97 | 9.4e-05 | Click! |
| PHOX2A | hg19_v2_chr11_-_71955210_71955227, hg19_v2_chr11_-_71952236_71952319 | -0.64 | 8.7e-02 | Click! |
| ALX4 | hg19_v2_chr11_-_44331679_44331716 | -0.57 | 1.4e-01 | Click! |
Activity profile of PRRX1_ALX4_PHOX2A motif
Sorted Z-values of PRRX1_ALX4_PHOX2A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PRRX1_ALX4_PHOX2A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_69865866 | 5.05 |
ENST00000354393.2 |
MYPN |
myopalladin |
| chr1_-_92371839 | 4.55 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr12_-_91546926 | 3.78 |
ENST00000550758.1 |
DCN |
decorin |
| chr2_-_145278475 | 3.57 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr13_+_102142296 | 3.07 |
ENST00000376162.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr5_-_111093759 | 2.79 |
ENST00000509979.1 ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP |
neuronal regeneration related protein |
| chr1_+_183774240 | 2.54 |
ENST00000360851.3 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr5_-_111312622 | 2.22 |
ENST00000395634.3 |
NREP |
neuronal regeneration related protein |
| chr1_-_232651312 | 1.90 |
ENST00000262861.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr3_-_195310802 | 1.89 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
| chr2_+_152214098 | 1.64 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr12_-_91573132 | 1.32 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr12_-_91573316 | 1.28 |
ENST00000393155.1 |
DCN |
decorin |
| chr12_+_59989918 | 1.20 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_-_145275228 | 1.19 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chrX_+_77166172 | 1.15 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr16_+_53133070 | 1.09 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr11_-_96076334 | 1.09 |
ENST00000524717.1 |
MAML2 |
mastermind-like 2 (Drosophila) |
| chr3_+_69985734 | 1.06 |
ENST00000314557.6 ENST00000394351.3 |
MITF |
microphthalmia-associated transcription factor |
| chr3_-_164914640 | 0.97 |
ENST00000241274.3 |
SLITRK3 |
SLIT and NTRK-like family, member 3 |
| chr5_+_140743859 | 0.89 |
ENST00000518069.1 |
PCDHGA5 |
protocadherin gamma subfamily A, 5 |
| chr4_+_160188889 | 0.88 |
ENST00000264431.4 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr8_-_122653630 | 0.87 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr7_+_90338712 | 0.85 |
ENST00000265741.3 ENST00000406263.1 |
CDK14 |
cyclin-dependent kinase 14 |
| chr12_-_52946923 | 0.84 |
ENST00000267119.5 |
KRT71 |
keratin 71 |
| chr20_+_31823792 | 0.83 |
ENST00000375413.4 ENST00000354297.4 ENST00000375422.2 |
BPIFA1 |
BPI fold containing family A, member 1 |
| chr12_+_18414446 | 0.83 |
ENST00000433979.1 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr6_-_154751629 | 0.74 |
ENST00000424998.1 |
CNKSR3 |
CNKSR family member 3 |
| chr10_+_89420706 | 0.73 |
ENST00000427144.2 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr12_-_91573249 | 0.68 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr14_+_95078714 | 0.67 |
ENST00000393078.3 ENST00000393080.4 ENST00000467132.1 |
SERPINA3 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr12_-_47219733 | 0.62 |
ENST00000547477.1 ENST00000447411.1 ENST00000266579.4 |
SLC38A4 |
solute carrier family 38, member 4 |
| chr3_+_12329397 | 0.61 |
ENST00000397015.2 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr19_+_50016610 | 0.61 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr6_+_34204642 | 0.58 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr1_-_241799232 | 0.55 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chr17_+_39240459 | 0.54 |
ENST00000391417.4 |
KRTAP4-7 |
keratin associated protein 4-7 |
| chr5_-_160279207 | 0.53 |
ENST00000327245.5 |
ATP10B |
ATPase, class V, type 10B |
| chr9_-_95244781 | 0.53 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr5_+_140762268 | 0.50 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr8_-_93107443 | 0.46 |
ENST00000360348.2 ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr17_-_5321549 | 0.45 |
ENST00000572809.1 |
NUP88 |
nucleoporin 88kDa |
| chr20_-_34025999 | 0.45 |
ENST00000374369.3 |
GDF5 |
growth differentiation factor 5 |
| chr4_-_83769996 | 0.44 |
ENST00000511338.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr2_+_54683419 | 0.44 |
ENST00000356805.4 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
| chr8_+_38677850 | 0.43 |
ENST00000518809.1 ENST00000520611.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr2_-_145277882 | 0.42 |
ENST00000465070.1 ENST00000444559.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr1_-_68698222 | 0.42 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chrX_+_9431324 | 0.41 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr1_-_68698197 | 0.40 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr3_+_69985792 | 0.39 |
ENST00000531774.1 |
MITF |
microphthalmia-associated transcription factor |
| chr8_-_57472154 | 0.39 |
ENST00000499425.1 ENST00000523664.1 ENST00000518943.1 ENST00000524338.1 |
LINC00968 |
long intergenic non-protein coding RNA 968 |
| chr2_-_175711133 | 0.38 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr3_-_186262166 | 0.38 |
ENST00000307944.5 |
CRYGS |
crystallin, gamma S |
| chr6_-_152639479 | 0.36 |
ENST00000356820.4 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr1_+_206557366 | 0.36 |
ENST00000414007.1 ENST00000419187.2 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
| chr11_-_63439013 | 0.35 |
ENST00000398868.3 |
ATL3 |
atlastin GTPase 3 |
| chr13_+_53602894 | 0.35 |
ENST00000219022.2 |
OLFM4 |
olfactomedin 4 |
| chr10_+_90672113 | 0.34 |
ENST00000371922.1 |
STAMBPL1 |
STAM binding protein-like 1 |
| chr8_-_93107696 | 0.34 |
ENST00000436581.2 ENST00000520583.1 ENST00000519061.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr8_-_26724784 | 0.33 |
ENST00000380573.3 |
ADRA1A |
adrenoceptor alpha 1A |
| chr7_+_90339169 | 0.32 |
ENST00000436577.2 |
CDK14 |
cyclin-dependent kinase 14 |
| chr15_+_51669444 | 0.32 |
ENST00000396399.2 |
GLDN |
gliomedin |
| chr15_-_52970820 | 0.29 |
ENST00000261844.7 ENST00000399202.4 ENST00000562135.1 |
FAM214A |
family with sequence similarity 214, member A |
| chr17_+_44588877 | 0.28 |
ENST00000576629.1 |
LRRC37A2 |
leucine rich repeat containing 37, member A2 |
| chr12_-_118796910 | 0.27 |
ENST00000541186.1 ENST00000539872.1 |
TAOK3 |
TAO kinase 3 |
| chr7_-_27169801 | 0.27 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr2_+_101437487 | 0.27 |
ENST00000427413.1 ENST00000542504.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr12_-_107380846 | 0.26 |
ENST00000548101.1 ENST00000550496.1 ENST00000552029.1 |
MTERFD3 |
MTERF domain containing 3 |
| chr2_+_228678550 | 0.25 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chr10_-_31146615 | 0.25 |
ENST00000444692.2 |
ZNF438 |
zinc finger protein 438 |
| chr20_-_18477862 | 0.24 |
ENST00000337227.4 |
RBBP9 |
retinoblastoma binding protein 9 |
| chr7_+_148287657 | 0.24 |
ENST00000307003.2 |
C7orf33 |
chromosome 7 open reading frame 33 |
| chr12_-_92539614 | 0.24 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr3_+_138340049 | 0.24 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr5_+_137203465 | 0.24 |
ENST00000239926.4 |
MYOT |
myotilin |
| chr6_-_107235331 | 0.24 |
ENST00000433965.1 ENST00000430094.1 |
RP1-60O19.1 |
RP1-60O19.1 |
| chr12_-_107380910 | 0.24 |
ENST00000392830.2 ENST00000240050.4 |
MTERFD3 |
MTERF domain containing 3 |
| chr5_+_140079919 | 0.23 |
ENST00000274712.3 |
ZMAT2 |
zinc finger, matrin-type 2 |
| chr8_+_77318769 | 0.23 |
ENST00000518732.1 |
RP11-706J10.1 |
long intergenic non-protein coding RNA 1111 |
| chr17_-_56082455 | 0.23 |
ENST00000578794.1 |
RP11-159D12.5 |
Uncharacterized protein |
| chr1_+_114471809 | 0.22 |
ENST00000426820.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr15_-_37393406 | 0.22 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr3_-_124774802 | 0.22 |
ENST00000311127.4 |
HEG1 |
heart development protein with EGF-like domains 1 |
| chr2_-_161350305 | 0.22 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr7_+_55980331 | 0.22 |
ENST00000429591.2 |
ZNF713 |
zinc finger protein 713 |
| chr5_-_142780280 | 0.22 |
ENST00000424646.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr11_+_12766583 | 0.22 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr4_+_165878100 | 0.21 |
ENST00000513876.2 |
FAM218A |
family with sequence similarity 218, member A |
| chr4_-_119759795 | 0.21 |
ENST00000419654.2 |
SEC24D |
SEC24 family member D |
| chr17_-_39623681 | 0.21 |
ENST00000225899.3 |
KRT32 |
keratin 32 |
| chr3_+_138340067 | 0.21 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr5_+_140571902 | 0.21 |
ENST00000239446.4 |
PCDHB10 |
protocadherin beta 10 |
| chr12_+_75784850 | 0.21 |
ENST00000550916.1 ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2 |
GLI pathogenesis-related 1 like 2 |
| chr6_+_153552455 | 0.20 |
ENST00000392385.2 |
AL590867.1 |
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr3_+_159943362 | 0.20 |
ENST00000326474.3 |
C3orf80 |
chromosome 3 open reading frame 80 |
| chr1_+_12916941 | 0.20 |
ENST00000240189.2 |
PRAMEF2 |
PRAME family member 2 |
| chr4_+_113568207 | 0.20 |
ENST00000511529.1 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chr11_-_82708435 | 0.20 |
ENST00000525117.1 ENST00000532548.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr10_-_29923893 | 0.20 |
ENST00000355867.4 |
SVIL |
supervillin |
| chr3_-_12587055 | 0.20 |
ENST00000564146.3 |
C3orf83 |
chromosome 3 open reading frame 83 |
| chr5_+_140514782 | 0.20 |
ENST00000231134.5 |
PCDHB5 |
protocadherin beta 5 |
| chr1_+_84630645 | 0.20 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr15_+_66874502 | 0.19 |
ENST00000558797.1 |
RP11-321F6.1 |
HCG2003567; Uncharacterized protein |
| chr3_-_157824292 | 0.19 |
ENST00000483851.2 |
SHOX2 |
short stature homeobox 2 |
| chr19_+_13842559 | 0.19 |
ENST00000586600.1 |
CCDC130 |
coiled-coil domain containing 130 |
| chr8_-_57123815 | 0.18 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr4_+_76649797 | 0.18 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr19_-_44388116 | 0.18 |
ENST00000587539.1 |
ZNF404 |
zinc finger protein 404 |
| chr1_+_52682052 | 0.18 |
ENST00000371591.1 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
| chr5_+_98109322 | 0.18 |
ENST00000513185.1 |
RGMB |
repulsive guidance molecule family member b |
| chr7_-_140482926 | 0.18 |
ENST00000496384.2 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr12_-_53601055 | 0.18 |
ENST00000552972.1 ENST00000422257.3 ENST00000267082.5 |
ITGB7 |
integrin, beta 7 |
| chr4_+_142558078 | 0.18 |
ENST00000529613.1 |
IL15 |
interleukin 15 |
| chr1_+_44679159 | 0.17 |
ENST00000315913.5 ENST00000372289.2 |
DMAP1 |
DNA methyltransferase 1 associated protein 1 |
| chr3_+_111393501 | 0.17 |
ENST00000393934.3 |
PLCXD2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr13_-_81801115 | 0.17 |
ENST00000567258.1 |
LINC00564 |
long intergenic non-protein coding RNA 564 |
| chr5_+_59783540 | 0.17 |
ENST00000515734.2 |
PART1 |
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr12_+_120105558 | 0.17 |
ENST00000229328.5 ENST00000541640.1 |
PRKAB1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr12_-_53601000 | 0.16 |
ENST00000338737.4 ENST00000549086.2 |
ITGB7 |
integrin, beta 7 |
| chr11_+_66276550 | 0.16 |
ENST00000419755.3 |
CTD-3074O7.11 |
Bardet-Biedl syndrome 1 protein |
| chr14_+_52164820 | 0.16 |
ENST00000554167.1 |
FRMD6 |
FERM domain containing 6 |
| chr10_+_13142225 | 0.16 |
ENST00000378747.3 |
OPTN |
optineurin |
| chr17_-_14683517 | 0.16 |
ENST00000379640.1 |
AC005863.1 |
AC005863.1 |
| chr17_-_71228357 | 0.16 |
ENST00000583024.1 ENST00000403627.3 ENST00000405159.3 ENST00000581110.1 |
FAM104A |
family with sequence similarity 104, member A |
| chr17_+_74463650 | 0.16 |
ENST00000392492.3 |
AANAT |
aralkylamine N-acetyltransferase |
| chr6_+_116691001 | 0.16 |
ENST00000537543.1 |
DSE |
dermatan sulfate epimerase |
| chr5_-_66492562 | 0.15 |
ENST00000256447.4 |
CD180 |
CD180 molecule |
| chrX_-_15332665 | 0.15 |
ENST00000537676.1 ENST00000344384.4 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr15_+_44719970 | 0.15 |
ENST00000558966.1 |
CTDSPL2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr14_+_88851874 | 0.15 |
ENST00000393545.4 ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7 |
spermatogenesis associated 7 |
| chr3_-_149293990 | 0.15 |
ENST00000472417.1 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr14_-_75536182 | 0.15 |
ENST00000555463.1 |
ACYP1 |
acylphosphatase 1, erythrocyte (common) type |
| chr7_+_138145076 | 0.15 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr3_+_40518599 | 0.14 |
ENST00000314686.5 ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619 |
zinc finger protein 619 |
| chr16_+_1728257 | 0.14 |
ENST00000248098.3 ENST00000562684.1 ENST00000561516.1 ENST00000382711.5 ENST00000566742.1 |
HN1L |
hematological and neurological expressed 1-like |
| chr3_-_18480260 | 0.14 |
ENST00000454909.2 |
SATB1 |
SATB homeobox 1 |
| chr5_+_68860949 | 0.14 |
ENST00000507595.1 |
GTF2H2C |
general transcription factor IIH, polypeptide 2C |
| chr5_-_70272055 | 0.14 |
ENST00000514857.2 |
NAIP |
NLR family, apoptosis inhibitory protein |
| chr10_-_90967063 | 0.14 |
ENST00000371852.2 |
CH25H |
cholesterol 25-hydroxylase |
| chr12_-_81992111 | 0.14 |
ENST00000443686.3 ENST00000407050.4 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr14_+_57671888 | 0.14 |
ENST00000391612.1 |
AL391152.1 |
AL391152.1 |
| chr10_+_35464513 | 0.14 |
ENST00000494479.1 ENST00000463314.1 ENST00000342105.3 ENST00000495301.1 ENST00000463960.1 |
CREM |
cAMP responsive element modulator |
| chr4_-_23735183 | 0.13 |
ENST00000507666.1 |
RP11-380P13.2 |
RP11-380P13.2 |
| chr1_+_114471972 | 0.13 |
ENST00000369559.4 ENST00000369554.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr14_+_21467414 | 0.13 |
ENST00000554422.1 ENST00000298681.4 |
SLC39A2 |
solute carrier family 39 (zinc transporter), member 2 |
| chr19_-_11456905 | 0.13 |
ENST00000588560.1 ENST00000592952.1 |
TMEM205 |
transmembrane protein 205 |
| chr5_+_133562095 | 0.13 |
ENST00000602919.1 |
CTD-2410N18.3 |
CTD-2410N18.3 |
| chr8_+_11961898 | 0.13 |
ENST00000400085.3 |
ZNF705D |
zinc finger protein 705D |
| chr17_-_46690839 | 0.13 |
ENST00000498634.2 |
HOXB8 |
homeobox B8 |
| chr9_-_95298314 | 0.13 |
ENST00000344604.5 ENST00000375540.1 |
ECM2 |
extracellular matrix protein 2, female organ and adipocyte specific |
| chr5_-_54468974 | 0.13 |
ENST00000381375.2 ENST00000296733.1 ENST00000322374.6 ENST00000334206.5 ENST00000331730.3 |
CDC20B |
cell division cycle 20B |
| chr7_+_23338819 | 0.13 |
ENST00000466681.1 |
MALSU1 |
mitochondrial assembly of ribosomal large subunit 1 |
| chr8_-_62602327 | 0.13 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr19_-_11456722 | 0.12 |
ENST00000354882.5 |
TMEM205 |
transmembrane protein 205 |
| chr15_-_75748115 | 0.12 |
ENST00000360439.4 |
SIN3A |
SIN3 transcription regulator family member A |
| chr9_+_2159850 | 0.12 |
ENST00000416751.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr6_+_31895254 | 0.12 |
ENST00000299367.5 ENST00000442278.2 |
C2 |
complement component 2 |
| chr1_+_158978768 | 0.12 |
ENST00000447473.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr12_-_10151773 | 0.12 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr10_+_134150835 | 0.12 |
ENST00000432555.2 |
LRRC27 |
leucine rich repeat containing 27 |
| chr15_+_75628394 | 0.12 |
ENST00000564815.1 ENST00000338995.6 |
COMMD4 |
COMM domain containing 4 |
| chr15_+_57511609 | 0.12 |
ENST00000543579.1 ENST00000537840.1 ENST00000343827.3 |
TCF12 |
transcription factor 12 |
| chr22_+_25615489 | 0.12 |
ENST00000398215.2 |
CRYBB2 |
crystallin, beta B2 |
| chr17_+_71228793 | 0.12 |
ENST00000426147.2 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chr1_-_165414414 | 0.11 |
ENST00000359842.5 |
RXRG |
retinoid X receptor, gamma |
| chr4_-_83765613 | 0.11 |
ENST00000503937.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr15_-_26874230 | 0.11 |
ENST00000400188.3 |
GABRB3 |
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr15_+_75628232 | 0.11 |
ENST00000267935.8 ENST00000567195.1 |
COMMD4 |
COMM domain containing 4 |
| chr2_-_58468437 | 0.11 |
ENST00000403676.1 ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL |
Fanconi anemia, complementation group L |
| chr6_+_132891461 | 0.11 |
ENST00000275198.1 |
TAAR6 |
trace amine associated receptor 6 |
| chr9_+_34652164 | 0.11 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chrX_-_48858630 | 0.11 |
ENST00000376425.3 ENST00000376444.3 |
GRIPAP1 |
GRIP1 associated protein 1 |
| chrX_-_48858667 | 0.11 |
ENST00000376423.4 ENST00000376441.1 |
GRIPAP1 |
GRIP1 associated protein 1 |
| chr2_-_172087824 | 0.11 |
ENST00000521943.1 |
TLK1 |
tousled-like kinase 1 |
| chr17_+_71228537 | 0.11 |
ENST00000577615.1 ENST00000585109.1 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chr9_-_123639600 | 0.11 |
ENST00000373896.3 |
PHF19 |
PHD finger protein 19 |
| chr1_+_154401791 | 0.11 |
ENST00000476006.1 |
IL6R |
interleukin 6 receptor |
| chr10_+_18689637 | 0.10 |
ENST00000377315.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chrX_+_16668278 | 0.10 |
ENST00000380200.3 |
S100G |
S100 calcium binding protein G |
| chr15_-_65407524 | 0.10 |
ENST00000559089.1 |
UBAP1L |
ubiquitin associated protein 1-like |
| chr5_+_169780485 | 0.10 |
ENST00000377360.4 |
KCNIP1 |
Kv channel interacting protein 1 |
| chr14_-_81425828 | 0.10 |
ENST00000555529.1 ENST00000556042.1 ENST00000556981.1 |
CEP128 |
centrosomal protein 128kDa |
| chr12_-_25150409 | 0.10 |
ENST00000549262.1 |
C12orf77 |
chromosome 12 open reading frame 77 |
| chr1_+_44679113 | 0.10 |
ENST00000361745.6 ENST00000446292.1 ENST00000440641.1 ENST00000436069.1 ENST00000437511.1 |
DMAP1 |
DNA methyltransferase 1 associated protein 1 |
| chr8_+_7353368 | 0.10 |
ENST00000355602.2 |
DEFB107B |
defensin, beta 107B |
| chr19_-_51920835 | 0.10 |
ENST00000442846.3 ENST00000530476.1 |
SIGLEC10 |
sialic acid binding Ig-like lectin 10 |
| chr19_-_11456872 | 0.10 |
ENST00000586218.1 |
TMEM205 |
transmembrane protein 205 |
| chr12_+_12878829 | 0.10 |
ENST00000326765.6 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr5_-_16738451 | 0.10 |
ENST00000274203.9 ENST00000515803.1 |
MYO10 |
myosin X |
| chr11_-_111649015 | 0.10 |
ENST00000529841.1 |
RP11-108O10.2 |
RP11-108O10.2 |
| chr10_+_13141585 | 0.09 |
ENST00000378764.2 |
OPTN |
optineurin |
| chr19_+_36630454 | 0.09 |
ENST00000246533.3 |
CAPNS1 |
calpain, small subunit 1 |
| chr12_-_43833515 | 0.09 |
ENST00000549670.1 ENST00000395541.2 |
ADAMTS20 |
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr21_+_48055527 | 0.09 |
ENST00000397638.2 ENST00000458387.2 ENST00000451211.2 ENST00000291705.6 ENST00000397637.1 ENST00000334494.4 ENST00000397628.1 ENST00000440086.1 |
PRMT2 |
protein arginine methyltransferase 2 |
| chr9_-_21305312 | 0.09 |
ENST00000259555.4 |
IFNA5 |
interferon, alpha 5 |
| chrX_+_35937843 | 0.09 |
ENST00000297866.5 |
CXorf22 |
chromosome X open reading frame 22 |
| chr12_-_10978957 | 0.09 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chrX_-_19817869 | 0.09 |
ENST00000379698.4 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr1_+_115572415 | 0.09 |
ENST00000256592.1 |
TSHB |
thyroid stimulating hormone, beta |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) response to luteinizing hormone(GO:0034699) |
| 0.9 | 5.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.6 | 1.9 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.4 | 6.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.4 | 1.1 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.3 | 0.9 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.3 | 0.9 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.3 | 5.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 0.8 | GO:1900191 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.2 | 0.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 0.6 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 1.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.5 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 1.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 0.6 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.3 | GO:0003099 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 0.3 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 0.3 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.6 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 0.5 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.4 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.2 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.1 | 0.2 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.4 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.9 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.9 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 1.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.8 | GO:0039694 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.6 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.9 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.2 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.0 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.5 | 7.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 1.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.3 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 5.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 2.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.4 | 4.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.4 | 1.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.3 | 1.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 0.7 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 0.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.9 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.2 | 5.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 0.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 5.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 6.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 1.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.3 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0097027 | anaphase-promoting complex binding(GO:0010997) ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 5.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |