Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for RAD21_SMC3
Z-value: 0.39
Transcription factors associated with RAD21_SMC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
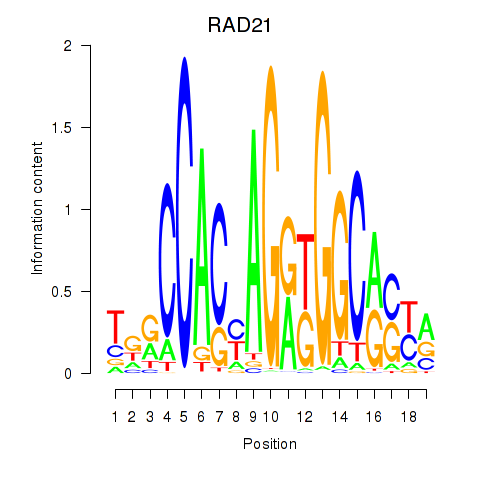
RAD21
|
ENSG00000164754.8 | RAD21 |
|
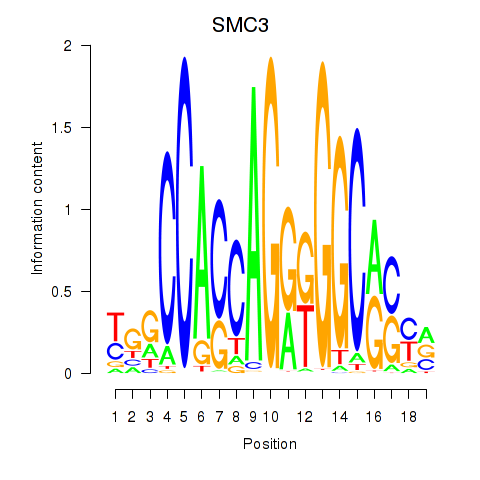
SMC3
|
ENSG00000108055.9 | SMC3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMC3 | hg19_v2_chr10_+_112327425_112327516 | 0.62 | 1.0e-01 | Click! |
| RAD21 | hg19_v2_chr8_-_117886955_117887105 | 0.04 | 9.3e-01 | Click! |
Activity profile of RAD21_SMC3 motif
Sorted Z-values of RAD21_SMC3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RAD21_SMC3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_114737472 | 2.19 |
ENST00000420161.1 |
AC110769.3 |
AC110769.3 |
| chr2_+_114737146 | 1.28 |
ENST00000412690.1 |
AC010982.1 |
AC010982.1 |
| chr7_+_89783689 | 1.15 |
ENST00000297205.2 |
STEAP1 |
six transmembrane epithelial antigen of the prostate 1 |
| chr13_+_31774073 | 1.05 |
ENST00000343307.4 |
B3GALTL |
beta 1,3-galactosyltransferase-like |
| chr6_+_138188551 | 0.86 |
ENST00000237289.4 ENST00000433680.1 |
TNFAIP3 |
tumor necrosis factor, alpha-induced protein 3 |
| chr15_+_45722727 | 0.86 |
ENST00000396650.2 ENST00000558435.1 ENST00000344300.3 |
C15orf48 |
chromosome 15 open reading frame 48 |
| chr5_-_139726181 | 0.85 |
ENST00000507104.1 ENST00000230990.6 |
HBEGF |
heparin-binding EGF-like growth factor |
| chr8_+_28480246 | 0.81 |
ENST00000523149.1 |
EXTL3 |
exostosin-like glycosyltransferase 3 |
| chr10_-_15413035 | 0.81 |
ENST00000378116.4 ENST00000455654.1 |
FAM171A1 |
family with sequence similarity 171, member A1 |
| chr19_-_36001113 | 0.81 |
ENST00000434389.1 |
DMKN |
dermokine |
| chr1_+_186344945 | 0.80 |
ENST00000419367.3 ENST00000287859.6 |
C1orf27 |
chromosome 1 open reading frame 27 |
| chr1_+_212208919 | 0.76 |
ENST00000366991.4 ENST00000542077.1 |
DTL |
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr10_-_76995675 | 0.74 |
ENST00000469299.1 |
COMTD1 |
catechol-O-methyltransferase domain containing 1 |
| chr19_-_36001286 | 0.73 |
ENST00000602679.1 ENST00000492341.2 ENST00000472252.2 ENST00000602781.1 ENST00000402589.2 ENST00000458071.1 ENST00000436012.1 ENST00000443640.1 ENST00000450261.1 ENST00000467637.1 ENST00000480502.1 ENST00000474928.1 ENST00000414866.2 ENST00000392206.2 ENST00000488892.1 |
DMKN |
dermokine |
| chr1_-_186344456 | 0.73 |
ENST00000367478.4 |
TPR |
translocated promoter region, nuclear basket protein |
| chr5_+_140254884 | 0.71 |
ENST00000398631.2 |
PCDHA12 |
protocadherin alpha 12 |
| chr10_-_76995769 | 0.71 |
ENST00000372538.3 |
COMTD1 |
catechol-O-methyltransferase domain containing 1 |
| chr15_-_34502197 | 0.71 |
ENST00000557877.1 |
KATNBL1 |
katanin p80 subunit B-like 1 |
| chr1_-_186344802 | 0.68 |
ENST00000451586.1 |
TPR |
translocated promoter region, nuclear basket protein |
| chr1_+_186344883 | 0.65 |
ENST00000367470.3 |
C1orf27 |
chromosome 1 open reading frame 27 |
| chr18_-_47721447 | 0.63 |
ENST00000285039.7 |
MYO5B |
myosin VB |
| chr7_-_22539771 | 0.61 |
ENST00000406890.2 ENST00000424363.1 |
STEAP1B |
STEAP family member 1B |
| chr1_-_159915386 | 0.58 |
ENST00000361509.3 ENST00000368094.1 |
IGSF9 |
immunoglobulin superfamily, member 9 |
| chr3_-_190040223 | 0.58 |
ENST00000295522.3 |
CLDN1 |
claudin 1 |
| chr1_+_92495528 | 0.55 |
ENST00000370383.4 |
EPHX4 |
epoxide hydrolase 4 |
| chr6_-_44225231 | 0.54 |
ENST00000538577.1 ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr19_+_58341656 | 0.53 |
ENST00000442832.4 ENST00000594901.1 |
ZNF587B |
zinc finger protein 587B |
| chr22_-_50050986 | 0.51 |
ENST00000405854.1 |
C22orf34 |
chromosome 22 open reading frame 34 |
| chr7_-_16840820 | 0.51 |
ENST00000450569.1 |
AGR2 |
anterior gradient 2 |
| chr11_+_60691924 | 0.49 |
ENST00000544065.1 ENST00000453848.2 ENST00000005286.4 |
TMEM132A |
transmembrane protein 132A |
| chr15_-_34659349 | 0.49 |
ENST00000314891.6 |
LPCAT4 |
lysophosphatidylcholine acyltransferase 4 |
| chr11_+_20409070 | 0.49 |
ENST00000331079.6 |
PRMT3 |
protein arginine methyltransferase 3 |
| chr1_-_94374946 | 0.49 |
ENST00000370238.3 |
GCLM |
glutamate-cysteine ligase, modifier subunit |
| chr10_-_79789291 | 0.47 |
ENST00000372371.3 |
POLR3A |
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr12_-_120763739 | 0.46 |
ENST00000549767.1 |
PLA2G1B |
phospholipase A2, group IB (pancreas) |
| chr4_+_56815102 | 0.46 |
ENST00000257287.4 |
CEP135 |
centrosomal protein 135kDa |
| chr18_-_21166841 | 0.45 |
ENST00000269228.5 |
NPC1 |
Niemann-Pick disease, type C1 |
| chr9_+_116917807 | 0.45 |
ENST00000356083.3 |
COL27A1 |
collagen, type XXVII, alpha 1 |
| chr8_+_67341239 | 0.44 |
ENST00000320270.2 |
RRS1 |
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr10_+_81107216 | 0.43 |
ENST00000394579.3 ENST00000225174.3 |
PPIF |
peptidylprolyl isomerase F |
| chr17_-_34890732 | 0.42 |
ENST00000268852.9 |
MYO19 |
myosin XIX |
| chr8_+_24772455 | 0.40 |
ENST00000433454.2 |
NEFM |
neurofilament, medium polypeptide |
| chr19_+_4791722 | 0.40 |
ENST00000269856.3 |
FEM1A |
fem-1 homolog a (C. elegans) |
| chr20_-_4721314 | 0.39 |
ENST00000418528.1 ENST00000326539.2 ENST00000423718.2 |
PRNT |
prion protein (testis specific) |
| chr8_-_145550571 | 0.39 |
ENST00000332324.4 |
DGAT1 |
diacylglycerol O-acyltransferase 1 |
| chr11_+_64216546 | 0.39 |
ENST00000316124.2 |
AP003774.4 |
HCG1652096, isoform CRA_a; Uncharacterized protein; cDNA FLJ37045 fis, clone BRACE2012185 |
| chrX_-_117119243 | 0.39 |
ENST00000539496.1 ENST00000469946.1 |
KLHL13 |
kelch-like family member 13 |
| chr3_+_172468472 | 0.39 |
ENST00000232458.5 ENST00000392692.3 |
ECT2 |
epithelial cell transforming sequence 2 oncogene |
| chr11_-_11643540 | 0.39 |
ENST00000227756.4 |
GALNT18 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18 |
| chr12_+_50898881 | 0.38 |
ENST00000301180.5 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr22_+_22735135 | 0.38 |
ENST00000390297.2 |
IGLV1-44 |
immunoglobulin lambda variable 1-44 |
| chr19_-_1876156 | 0.38 |
ENST00000565797.1 |
CTB-31O20.2 |
CTB-31O20.2 |
| chr17_-_61523622 | 0.38 |
ENST00000448884.2 ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561 |
cytochrome b561 |
| chr3_+_50192499 | 0.37 |
ENST00000413852.1 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr15_+_75074385 | 0.37 |
ENST00000220003.9 |
CSK |
c-src tyrosine kinase |
| chr13_-_37573432 | 0.37 |
ENST00000413537.2 ENST00000443765.1 ENST00000239891.3 |
ALG5 |
ALG5, dolichyl-phosphate beta-glucosyltransferase |
| chr4_-_41884620 | 0.37 |
ENST00000504870.1 |
LINC00682 |
long intergenic non-protein coding RNA 682 |
| chr3_+_172468505 | 0.36 |
ENST00000427830.1 ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2 |
epithelial cell transforming sequence 2 oncogene |
| chr1_-_40367530 | 0.36 |
ENST00000372816.2 ENST00000372815.1 |
MYCL |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr17_-_34890759 | 0.36 |
ENST00000431794.3 |
MYO19 |
myosin XIX |
| chr10_-_103603523 | 0.35 |
ENST00000370046.1 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr16_-_88851618 | 0.35 |
ENST00000301015.9 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
| chr5_+_175875349 | 0.35 |
ENST00000261942.6 |
FAF2 |
Fas associated factor family member 2 |
| chr1_+_15480197 | 0.35 |
ENST00000400796.3 ENST00000434578.2 ENST00000376008.2 |
TMEM51 |
transmembrane protein 51 |
| chr5_+_145826867 | 0.35 |
ENST00000296702.5 ENST00000394421.2 |
TCERG1 |
transcription elongation regulator 1 |
| chr2_+_69240415 | 0.34 |
ENST00000409829.3 |
ANTXR1 |
anthrax toxin receptor 1 |
| chr11_-_104480019 | 0.34 |
ENST00000536529.1 ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1 |
RP11-886D15.1 |
| chr6_-_32806506 | 0.34 |
ENST00000374897.2 ENST00000452392.2 |
TAP2 TAP2 |
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr12_+_13197218 | 0.33 |
ENST00000197268.8 |
KIAA1467 |
KIAA1467 |
| chr17_+_7621045 | 0.33 |
ENST00000570791.1 |
DNAH2 |
dynein, axonemal, heavy chain 2 |
| chr19_-_58400148 | 0.33 |
ENST00000595048.1 ENST00000600634.1 ENST00000595295.1 ENST00000596604.1 ENST00000597342.1 ENST00000597807.1 |
ZNF814 |
zinc finger protein 814 |
| chr5_+_140165876 | 0.33 |
ENST00000504120.2 ENST00000394633.3 ENST00000378133.3 |
PCDHA1 |
protocadherin alpha 1 |
| chr8_+_101170563 | 0.32 |
ENST00000520508.1 ENST00000388798.2 |
SPAG1 |
sperm associated antigen 1 |
| chr6_+_151186554 | 0.32 |
ENST00000367321.3 ENST00000367307.4 |
MTHFD1L |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr11_-_113644491 | 0.32 |
ENST00000200135.3 |
ZW10 |
zw10 kinetochore protein |
| chr16_-_66864806 | 0.32 |
ENST00000566336.1 ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
| chr7_+_156742399 | 0.31 |
ENST00000275820.3 |
NOM1 |
nucleolar protein with MIF4G domain 1 |
| chr7_+_150521691 | 0.31 |
ENST00000493429.1 |
AOC1 |
amine oxidase, copper containing 1 |
| chr17_+_38137073 | 0.31 |
ENST00000541736.1 |
PSMD3 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 |
| chr5_+_140201183 | 0.31 |
ENST00000529619.1 ENST00000529859.1 ENST00000378126.3 |
PCDHA5 |
protocadherin alpha 5 |
| chr12_+_110338063 | 0.30 |
ENST00000405876.4 |
TCHP |
trichoplein, keratin filament binding |
| chrX_+_49969405 | 0.30 |
ENST00000376042.1 |
CCNB3 |
cyclin B3 |
| chr3_-_196756646 | 0.30 |
ENST00000439320.1 ENST00000296351.4 ENST00000296350.5 |
MFI2 |
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
| chr8_-_145669791 | 0.30 |
ENST00000409379.3 |
TONSL |
tonsoku-like, DNA repair protein |
| chr2_+_11273179 | 0.30 |
ENST00000381585.3 ENST00000405022.3 |
C2orf50 |
chromosome 2 open reading frame 50 |
| chr20_+_55205825 | 0.30 |
ENST00000544508.1 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr2_+_220492287 | 0.29 |
ENST00000273063.6 ENST00000373762.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr10_+_13203543 | 0.29 |
ENST00000378714.3 ENST00000479669.1 ENST00000484800.2 |
MCM10 |
minichromosome maintenance complex component 10 |
| chr6_-_11044509 | 0.29 |
ENST00000354666.3 |
ELOVL2 |
ELOVL fatty acid elongase 2 |
| chr4_+_56814968 | 0.29 |
ENST00000422247.2 |
CEP135 |
centrosomal protein 135kDa |
| chr19_-_49496557 | 0.29 |
ENST00000323798.3 ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1 |
glycogen synthase 1 (muscle) |
| chr14_-_54908043 | 0.28 |
ENST00000556113.1 ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
| chr13_+_33160553 | 0.28 |
ENST00000315596.10 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr15_+_92396920 | 0.28 |
ENST00000318445.6 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
| chr18_+_60382672 | 0.28 |
ENST00000400316.4 ENST00000262719.5 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
| chr1_+_18081804 | 0.28 |
ENST00000375406.1 |
ACTL8 |
actin-like 8 |
| chr12_-_102224704 | 0.28 |
ENST00000299314.7 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr8_+_101170257 | 0.28 |
ENST00000251809.3 |
SPAG1 |
sperm associated antigen 1 |
| chr17_-_27507395 | 0.28 |
ENST00000354329.4 ENST00000527372.1 |
MYO18A |
myosin XVIIIA |
| chr10_-_6019984 | 0.28 |
ENST00000525219.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr22_+_22901750 | 0.28 |
ENST00000407120.1 |
LL22NC03-63E9.3 |
Uncharacterized protein |
| chr10_-_96122682 | 0.28 |
ENST00000371361.3 |
NOC3L |
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr10_-_101769617 | 0.27 |
ENST00000324109.4 ENST00000342239.3 |
DNMBP |
dynamin binding protein |
| chr2_+_220492373 | 0.27 |
ENST00000317151.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr11_-_119599794 | 0.27 |
ENST00000264025.3 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr16_+_22308717 | 0.27 |
ENST00000299853.5 ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E |
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr16_-_23568651 | 0.27 |
ENST00000563232.1 ENST00000563459.1 ENST00000449606.1 |
EARS2 |
glutamyl-tRNA synthetase 2, mitochondrial |
| chr17_-_72919317 | 0.27 |
ENST00000319642.1 |
USH1G |
Usher syndrome 1G (autosomal recessive) |
| chr17_-_27507377 | 0.26 |
ENST00000531253.1 |
MYO18A |
myosin XVIIIA |
| chr14_+_22356029 | 0.26 |
ENST00000390437.2 |
TRAV12-2 |
T cell receptor alpha variable 12-2 |
| chr1_+_151129103 | 0.26 |
ENST00000368910.3 |
TNFAIP8L2 |
tumor necrosis factor, alpha-induced protein 8-like 2 |
| chr12_+_110338323 | 0.26 |
ENST00000312777.5 ENST00000536408.2 |
TCHP |
trichoplein, keratin filament binding |
| chr21_-_36262032 | 0.26 |
ENST00000325074.5 ENST00000399237.2 |
RUNX1 |
runt-related transcription factor 1 |
| chr6_-_139308777 | 0.26 |
ENST00000529597.1 ENST00000415951.2 ENST00000367663.4 ENST00000409812.2 |
REPS1 |
RALBP1 associated Eps domain containing 1 |
| chr14_+_22217447 | 0.26 |
ENST00000390427.3 |
TRAV5 |
T cell receptor alpha variable 5 |
| chr6_+_155054459 | 0.26 |
ENST00000367178.3 ENST00000417268.1 ENST00000367186.4 |
SCAF8 |
SR-related CTD-associated factor 8 |
| chr22_-_43583079 | 0.26 |
ENST00000216129.6 |
TTLL12 |
tubulin tyrosine ligase-like family, member 12 |
| chr11_+_85566422 | 0.25 |
ENST00000342404.3 |
CCDC83 |
coiled-coil domain containing 83 |
| chr12_-_51663959 | 0.25 |
ENST00000604188.1 ENST00000398453.3 |
SMAGP |
small cell adhesion glycoprotein |
| chr9_-_35096545 | 0.25 |
ENST00000378617.3 ENST00000341666.3 ENST00000361778.2 |
PIGO |
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr1_-_6614565 | 0.25 |
ENST00000377705.5 |
NOL9 |
nucleolar protein 9 |
| chr8_+_42948641 | 0.25 |
ENST00000518991.1 ENST00000331373.5 |
POMK |
protein-O-mannose kinase |
| chr22_-_50051151 | 0.25 |
ENST00000400023.1 ENST00000444628.1 |
C22orf34 |
chromosome 22 open reading frame 34 |
| chr5_+_137688285 | 0.24 |
ENST00000314358.5 |
KDM3B |
lysine (K)-specific demethylase 3B |
| chr15_-_44487408 | 0.24 |
ENST00000402883.1 ENST00000417257.1 |
FRMD5 |
FERM domain containing 5 |
| chr16_+_16326352 | 0.24 |
ENST00000399336.4 ENST00000263012.6 ENST00000538468.1 |
NOMO3 |
NODAL modulator 3 |
| chr12_-_121410095 | 0.24 |
ENST00000539163.1 |
AC079602.1 |
AC079602.1 |
| chr5_+_150827143 | 0.24 |
ENST00000243389.3 ENST00000517945.1 ENST00000521925.1 |
SLC36A1 |
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr4_-_41884582 | 0.24 |
ENST00000499082.2 |
LINC00682 |
long intergenic non-protein coding RNA 682 |
| chr11_+_7110165 | 0.24 |
ENST00000306904.5 |
RBMXL2 |
RNA binding motif protein, X-linked-like 2 |
| chr12_-_51663728 | 0.24 |
ENST00000603864.1 ENST00000605426.1 |
SMAGP |
small cell adhesion glycoprotein |
| chr5_-_140998616 | 0.24 |
ENST00000389054.3 ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1 |
diaphanous-related formin 1 |
| chr17_+_17584763 | 0.24 |
ENST00000353383.1 |
RAI1 |
retinoic acid induced 1 |
| chr19_-_36004543 | 0.24 |
ENST00000339686.3 ENST00000447113.2 ENST00000440396.1 |
DMKN |
dermokine |
| chr11_+_66025938 | 0.24 |
ENST00000394066.2 |
KLC2 |
kinesin light chain 2 |
| chr17_+_65713925 | 0.24 |
ENST00000253247.4 |
NOL11 |
nucleolar protein 11 |
| chr12_-_95510743 | 0.23 |
ENST00000551521.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr19_+_10123925 | 0.23 |
ENST00000591589.1 ENST00000171214.1 |
RDH8 |
retinol dehydrogenase 8 (all-trans) |
| chr2_+_90108504 | 0.23 |
ENST00000390271.2 |
IGKV6D-41 |
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr16_-_18573396 | 0.23 |
ENST00000543392.1 ENST00000381474.3 ENST00000330537.6 |
NOMO2 |
NODAL modulator 2 |
| chr6_-_139309378 | 0.23 |
ENST00000450536.2 ENST00000258062.5 |
REPS1 |
RALBP1 associated Eps domain containing 1 |
| chr5_+_140220769 | 0.23 |
ENST00000531613.1 ENST00000378123.3 |
PCDHA8 |
protocadherin alpha 8 |
| chr22_+_22764088 | 0.23 |
ENST00000390299.2 |
IGLV1-40 |
immunoglobulin lambda variable 1-40 |
| chr8_+_22446763 | 0.23 |
ENST00000450780.2 ENST00000430850.2 ENST00000447849.1 |
AC037459.4 |
Uncharacterized protein |
| chr20_+_30102231 | 0.23 |
ENST00000335574.5 ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13 |
histocompatibility (minor) 13 |
| chr17_-_35716019 | 0.22 |
ENST00000591148.1 ENST00000394406.2 ENST00000394403.1 |
ACACA |
acetyl-CoA carboxylase alpha |
| chr7_-_91509986 | 0.22 |
ENST00000456229.1 ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF |
mitochondrial transcription termination factor |
| chr7_+_112120908 | 0.22 |
ENST00000439068.2 ENST00000312849.4 ENST00000429049.1 |
LSMEM1 |
leucine-rich single-pass membrane protein 1 |
| chr17_+_7591747 | 0.22 |
ENST00000534050.1 |
WRAP53 |
WD repeat containing, antisense to TP53 |
| chr1_-_156390128 | 0.22 |
ENST00000368242.3 |
C1orf61 |
chromosome 1 open reading frame 61 |
| chr2_+_102972363 | 0.22 |
ENST00000409599.1 |
IL18R1 |
interleukin 18 receptor 1 |
| chr17_+_65714018 | 0.22 |
ENST00000581106.1 ENST00000535137.1 |
NOL11 |
nucleolar protein 11 |
| chr17_+_38137050 | 0.22 |
ENST00000264639.4 |
PSMD3 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 |
| chr17_+_7591639 | 0.21 |
ENST00000396463.2 |
WRAP53 |
WD repeat containing, antisense to TP53 |
| chr16_+_1756162 | 0.21 |
ENST00000250894.4 ENST00000356010.5 |
MAPK8IP3 |
mitogen-activated protein kinase 8 interacting protein 3 |
| chr11_+_118826999 | 0.21 |
ENST00000264031.2 |
UPK2 |
uroplakin 2 |
| chr16_+_71392616 | 0.21 |
ENST00000349553.5 ENST00000302628.4 ENST00000562305.1 |
CALB2 |
calbindin 2 |
| chr11_+_46368975 | 0.21 |
ENST00000527911.1 |
DGKZ |
diacylglycerol kinase, zeta |
| chr3_+_141594966 | 0.21 |
ENST00000475483.1 |
ATP1B3 |
ATPase, Na+/K+ transporting, beta 3 polypeptide |
| chr14_+_23305760 | 0.21 |
ENST00000311852.6 |
MMP14 |
matrix metallopeptidase 14 (membrane-inserted) |
| chr2_+_69240511 | 0.21 |
ENST00000409349.3 |
ANTXR1 |
anthrax toxin receptor 1 |
| chr10_+_104154229 | 0.21 |
ENST00000428099.1 ENST00000369966.3 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr11_-_117103208 | 0.20 |
ENST00000320934.3 ENST00000530269.1 |
PCSK7 |
proprotein convertase subtilisin/kexin type 7 |
| chr4_-_128887069 | 0.20 |
ENST00000541133.1 ENST00000296468.3 ENST00000513559.1 |
MFSD8 |
major facilitator superfamily domain containing 8 |
| chr10_+_103892787 | 0.20 |
ENST00000278070.2 ENST00000413464.2 |
PPRC1 |
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr5_-_140998481 | 0.20 |
ENST00000518047.1 |
DIAPH1 |
diaphanous-related formin 1 |
| chr1_-_6761855 | 0.20 |
ENST00000426784.1 ENST00000377573.5 ENST00000377577.5 ENST00000294401.7 |
DNAJC11 |
DnaJ (Hsp40) homolog, subfamily C, member 11 |
| chr20_+_43992094 | 0.20 |
ENST00000453003.1 |
SYS1 |
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr14_+_22771851 | 0.20 |
ENST00000390466.1 |
TRAV39 |
T cell receptor alpha variable 39 |
| chr6_-_136871957 | 0.20 |
ENST00000354570.3 |
MAP7 |
microtubule-associated protein 7 |
| chr11_+_46369077 | 0.20 |
ENST00000456247.2 ENST00000421244.2 ENST00000318201.8 |
DGKZ |
diacylglycerol kinase, zeta |
| chr17_+_41561317 | 0.20 |
ENST00000540306.1 ENST00000262415.3 ENST00000605777.1 |
DHX8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr18_+_43753974 | 0.19 |
ENST00000282059.6 ENST00000321319.6 |
C18orf25 |
chromosome 18 open reading frame 25 |
| chr3_-_183735731 | 0.19 |
ENST00000334444.6 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr3_-_128369643 | 0.19 |
ENST00000296255.3 |
RPN1 |
ribophorin I |
| chr3_-_64009658 | 0.19 |
ENST00000394431.2 |
PSMD6 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr10_-_6622258 | 0.19 |
ENST00000263125.5 |
PRKCQ |
protein kinase C, theta |
| chr12_+_49372251 | 0.19 |
ENST00000293549.3 |
WNT1 |
wingless-type MMTV integration site family, member 1 |
| chr5_-_141257954 | 0.19 |
ENST00000456271.1 ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1 |
protocadherin 1 |
| chr3_+_140950612 | 0.19 |
ENST00000286353.4 ENST00000502783.1 ENST00000393010.2 ENST00000514680.1 |
ACPL2 |
acid phosphatase-like 2 |
| chr10_-_6622201 | 0.19 |
ENST00000539722.1 ENST00000397176.2 |
PRKCQ |
protein kinase C, theta |
| chr1_+_29063119 | 0.19 |
ENST00000474884.1 ENST00000542507.1 |
YTHDF2 |
YTH domain family, member 2 |
| chr11_+_46368956 | 0.19 |
ENST00000543978.1 |
DGKZ |
diacylglycerol kinase, zeta |
| chr1_-_211307404 | 0.18 |
ENST00000367007.4 |
KCNH1 |
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr3_-_50340996 | 0.18 |
ENST00000266031.4 ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1 |
hyaluronoglucosaminidase 1 |
| chr6_+_35310391 | 0.18 |
ENST00000337400.2 ENST00000311565.4 ENST00000540939.1 |
PPARD |
peroxisome proliferator-activated receptor delta |
| chr16_-_49315731 | 0.18 |
ENST00000219197.6 |
CBLN1 |
cerebellin 1 precursor |
| chrX_+_47053208 | 0.18 |
ENST00000442035.1 ENST00000457753.1 ENST00000335972.6 |
UBA1 |
ubiquitin-like modifier activating enzyme 1 |
| chr9_+_138594018 | 0.18 |
ENST00000487664.1 ENST00000298480.5 ENST00000371757.2 |
KCNT1 |
potassium channel, subfamily T, member 1 |
| chr14_+_22309368 | 0.18 |
ENST00000390433.1 |
TRAV12-1 |
T cell receptor alpha variable 12-1 |
| chr12_-_102224457 | 0.18 |
ENST00000549165.1 ENST00000549940.1 ENST00000392919.4 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr6_+_25279651 | 0.18 |
ENST00000329474.6 |
LRRC16A |
leucine rich repeat containing 16A |
| chr11_+_73675873 | 0.18 |
ENST00000537753.1 ENST00000542350.1 |
DNAJB13 |
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr1_-_211307315 | 0.18 |
ENST00000271751.4 |
KCNH1 |
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr19_+_47852538 | 0.18 |
ENST00000328771.4 |
DHX34 |
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
| chrX_-_103268259 | 0.17 |
ENST00000217926.5 |
H2BFWT |
H2B histone family, member W, testis-specific |
| chr4_+_7194247 | 0.17 |
ENST00000507866.2 |
SORCS2 |
sortilin-related VPS10 domain containing receptor 2 |
| chr17_-_33469299 | 0.17 |
ENST00000586869.1 ENST00000360831.5 ENST00000442241.4 |
NLE1 |
notchless homolog 1 (Drosophila) |
| chr11_-_130786400 | 0.17 |
ENST00000265909.4 |
SNX19 |
sorting nexin 19 |
| chr4_-_171011323 | 0.17 |
ENST00000509167.1 ENST00000353187.2 ENST00000507375.1 ENST00000515480.1 |
AADAT |
aminoadipate aminotransferase |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0045799 | mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.3 | 1.8 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 0.6 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.3 | 0.9 | GO:0071947 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) negative regulation of osteoclast proliferation(GO:0090291) |
| 0.2 | 0.9 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.2 | 0.7 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.2 | 1.8 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 0.5 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.2 | 0.5 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.2 | 0.5 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.2 | 0.5 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.4 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.4 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.4 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.4 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 | 0.5 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.6 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.3 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.4 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.3 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.5 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.5 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 0.3 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.5 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.5 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.5 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.4 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.5 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.2 | GO:0061054 | Spemann organizer formation(GO:0060061) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.1 | 0.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.3 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.1 | 0.2 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.5 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.2 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.4 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.2 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0035981 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.3 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.3 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.7 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.5 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.3 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.9 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 1.1 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.3 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.3 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.0 | 0.2 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.1 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.2 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.5 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:1904760 | peptidyl-serine ADP-ribosylation(GO:0018312) myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.5 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.2 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.4 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.3 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.6 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.7 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.3 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.1 | GO:0002501 | peptide antigen assembly with MHC protein complex(GO:0002501) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:1901165 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0070671 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.8 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 2.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.3 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.2 | 0.5 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 0.7 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.3 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 1.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.5 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.2 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.1 | 0.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.3 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 1.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.5 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.4 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 0.5 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 0.5 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.4 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 0.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.5 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.3 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.5 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.1 | 0.4 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.3 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.3 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 0.4 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.7 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.4 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.3 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.5 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.2 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 1.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.5 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |