Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
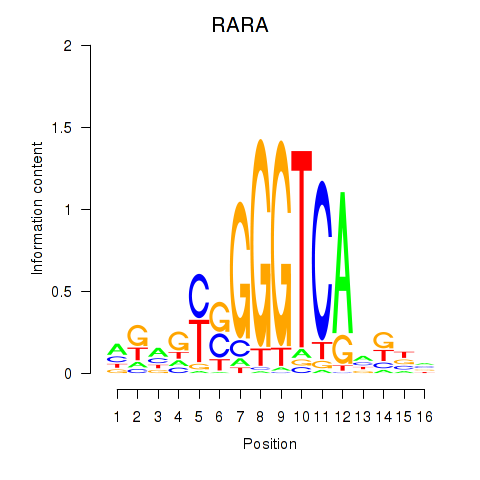
Results for RARA
Z-value: 0.44
Transcription factors associated with RARA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARA
|
ENSG00000131759.13 | RARA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARA | hg19_v2_chr17_+_38474489_38474548 | 0.72 | 4.5e-02 | Click! |
Activity profile of RARA motif
Sorted Z-values of RARA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RARA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_21529811 | 0.51 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr6_+_36097992 | 0.41 |
ENST00000211287.4 |
MAPK13 |
mitogen-activated protein kinase 13 |
| chr8_-_57233103 | 0.40 |
ENST00000303749.3 ENST00000522671.1 |
SDR16C5 |
short chain dehydrogenase/reductase family 16C, member 5 |
| chr3_-_69435224 | 0.37 |
ENST00000398540.3 |
FRMD4B |
FERM domain containing 4B |
| chr19_-_6720686 | 0.37 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr6_+_54711533 | 0.35 |
ENST00000306858.7 |
FAM83B |
family with sequence similarity 83, member B |
| chr2_-_113594279 | 0.31 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr6_+_7541845 | 0.31 |
ENST00000418664.2 |
DSP |
desmoplakin |
| chr17_-_34122596 | 0.30 |
ENST00000250144.8 |
MMP28 |
matrix metallopeptidase 28 |
| chr2_+_189157498 | 0.30 |
ENST00000359135.3 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr19_-_6767516 | 0.29 |
ENST00000245908.6 |
SH2D3A |
SH2 domain containing 3A |
| chr11_+_6866883 | 0.29 |
ENST00000299454.4 ENST00000379831.2 |
OR10A5 |
olfactory receptor, family 10, subfamily A, member 5 |
| chr19_-_6767431 | 0.27 |
ENST00000437152.3 ENST00000597687.1 |
SH2D3A |
SH2 domain containing 3A |
| chr8_-_57232656 | 0.27 |
ENST00000396721.2 |
SDR16C5 |
short chain dehydrogenase/reductase family 16C, member 5 |
| chr22_+_45072925 | 0.26 |
ENST00000006251.7 |
PRR5 |
proline rich 5 (renal) |
| chr22_+_45072958 | 0.26 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr2_+_189157536 | 0.26 |
ENST00000409580.1 ENST00000409637.3 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr18_+_19749386 | 0.25 |
ENST00000269216.3 |
GATA6 |
GATA binding protein 6 |
| chr11_+_45944190 | 0.25 |
ENST00000401752.1 ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B |
glycosyltransferase-like 1B |
| chr10_-_135090360 | 0.25 |
ENST00000486609.1 ENST00000445355.3 ENST00000485491.2 |
ADAM8 |
ADAM metallopeptidase domain 8 |
| chr19_-_19739007 | 0.25 |
ENST00000586703.1 ENST00000591042.1 ENST00000407877.3 |
LPAR2 |
lysophosphatidic acid receptor 2 |
| chr10_-_135090338 | 0.24 |
ENST00000415217.3 |
ADAM8 |
ADAM metallopeptidase domain 8 |
| chr2_-_235405679 | 0.24 |
ENST00000390645.2 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr1_-_161008697 | 0.24 |
ENST00000318289.10 ENST00000368023.3 ENST00000368024.1 ENST00000423014.2 |
TSTD1 |
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr11_+_62649158 | 0.23 |
ENST00000539891.1 ENST00000536981.1 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr16_-_85784557 | 0.22 |
ENST00000602675.1 |
C16orf74 |
chromosome 16 open reading frame 74 |
| chr1_-_156675368 | 0.22 |
ENST00000368222.3 |
CRABP2 |
cellular retinoic acid binding protein 2 |
| chr1_+_62208091 | 0.21 |
ENST00000316485.6 ENST00000371158.2 |
INADL |
InaD-like (Drosophila) |
| chr20_+_20348740 | 0.21 |
ENST00000310227.1 |
INSM1 |
insulinoma-associated 1 |
| chr1_+_233463507 | 0.21 |
ENST00000366623.3 ENST00000366624.3 |
MLK4 |
Mitogen-activated protein kinase kinase kinase MLK4 |
| chr1_-_156786530 | 0.20 |
ENST00000368198.3 |
SH2D2A |
SH2 domain containing 2A |
| chr19_-_1174226 | 0.20 |
ENST00000587024.1 ENST00000361757.3 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
| chr17_+_78075361 | 0.20 |
ENST00000577106.1 ENST00000390015.3 |
GAA |
glucosidase, alpha; acid |
| chr1_-_156786634 | 0.20 |
ENST00000392306.2 ENST00000368199.3 |
SH2D2A |
SH2 domain containing 2A |
| chr5_-_16509101 | 0.18 |
ENST00000399793.2 |
FAM134B |
family with sequence similarity 134, member B |
| chr2_+_220492287 | 0.18 |
ENST00000273063.6 ENST00000373762.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr12_-_85306562 | 0.17 |
ENST00000551612.1 ENST00000450363.3 ENST00000552192.1 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr2_+_220492116 | 0.17 |
ENST00000373760.2 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr22_+_50624323 | 0.17 |
ENST00000380909.4 ENST00000303434.4 |
TRABD |
TraB domain containing |
| chr1_+_101702417 | 0.16 |
ENST00000305352.6 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
| chr22_-_20255212 | 0.16 |
ENST00000416372.1 |
RTN4R |
reticulon 4 receptor |
| chr2_+_220492373 | 0.16 |
ENST00000317151.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr16_+_4838393 | 0.15 |
ENST00000589721.1 |
SMIM22 |
small integral membrane protein 22 |
| chr14_+_68086515 | 0.15 |
ENST00000261783.3 |
ARG2 |
arginase 2 |
| chr14_-_21493884 | 0.15 |
ENST00000556974.1 ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2 |
NDRG family member 2 |
| chr8_-_494824 | 0.15 |
ENST00000427263.2 ENST00000324079.6 |
TDRP |
testis development related protein |
| chr16_+_577697 | 0.14 |
ENST00000562370.1 ENST00000568988.1 ENST00000219611.2 |
CAPN15 |
calpain 15 |
| chr6_+_53659746 | 0.14 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr16_+_3115378 | 0.14 |
ENST00000529550.1 ENST00000551122.1 ENST00000525643.2 ENST00000548807.1 ENST00000528163.2 |
IL32 |
interleukin 32 |
| chr16_+_4838412 | 0.14 |
ENST00000589327.1 |
SMIM22 |
small integral membrane protein 22 |
| chr10_-_131762105 | 0.14 |
ENST00000368648.3 ENST00000355311.5 |
EBF3 |
early B-cell factor 3 |
| chr17_-_4463856 | 0.14 |
ENST00000574584.1 ENST00000381550.3 ENST00000301395.3 |
GGT6 |
gamma-glutamyltransferase 6 |
| chr4_-_90758118 | 0.13 |
ENST00000420646.2 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr22_-_21387127 | 0.13 |
ENST00000426145.1 |
SLC7A4 |
solute carrier family 7, member 4 |
| chr22_+_38302285 | 0.13 |
ENST00000215957.6 |
MICALL1 |
MICAL-like 1 |
| chr6_-_137365402 | 0.13 |
ENST00000541547.1 |
IL20RA |
interleukin 20 receptor, alpha |
| chr11_+_59480899 | 0.12 |
ENST00000300150.7 |
STX3 |
syntaxin 3 |
| chr15_+_22382382 | 0.12 |
ENST00000328795.4 |
OR4N4 |
olfactory receptor, family 4, subfamily N, member 4 |
| chr8_-_144815966 | 0.12 |
ENST00000388913.3 |
FAM83H |
family with sequence similarity 83, member H |
| chr3_-_112218205 | 0.12 |
ENST00000383680.4 |
BTLA |
B and T lymphocyte associated |
| chr22_-_29137771 | 0.12 |
ENST00000439200.1 ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2 |
checkpoint kinase 2 |
| chr3_+_13590636 | 0.12 |
ENST00000295760.7 |
FBLN2 |
fibulin 2 |
| chr10_-_103880209 | 0.12 |
ENST00000425280.1 |
LDB1 |
LIM domain binding 1 |
| chr14_+_24641062 | 0.11 |
ENST00000311457.3 ENST00000557806.1 ENST00000559919.1 |
REC8 |
REC8 meiotic recombination protein |
| chr18_+_44526786 | 0.11 |
ENST00000245121.5 ENST00000356157.7 |
KATNAL2 |
katanin p60 subunit A-like 2 |
| chr13_+_25670268 | 0.11 |
ENST00000281589.3 |
PABPC3 |
poly(A) binding protein, cytoplasmic 3 |
| chr12_+_133066137 | 0.11 |
ENST00000434748.2 |
FBRSL1 |
fibrosin-like 1 |
| chr15_+_74908147 | 0.11 |
ENST00000568139.1 ENST00000563297.1 ENST00000568488.1 ENST00000352989.5 ENST00000348245.3 |
CLK3 |
CDC-like kinase 3 |
| chr11_+_1856034 | 0.11 |
ENST00000341958.3 |
SYT8 |
synaptotagmin VIII |
| chr19_+_7701985 | 0.11 |
ENST00000595950.1 ENST00000441779.2 ENST00000221283.5 ENST00000414284.2 |
STXBP2 |
syntaxin binding protein 2 |
| chrX_+_99899180 | 0.10 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr16_+_71660052 | 0.10 |
ENST00000567566.1 |
MARVELD3 |
MARVEL domain containing 3 |
| chr14_-_91526462 | 0.10 |
ENST00000536315.2 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr17_+_78075498 | 0.10 |
ENST00000302262.3 |
GAA |
glucosidase, alpha; acid |
| chr11_+_1855645 | 0.10 |
ENST00000381968.3 ENST00000381978.3 |
SYT8 |
synaptotagmin VIII |
| chr15_-_34659349 | 0.10 |
ENST00000314891.6 |
LPCAT4 |
lysophosphatidylcholine acyltransferase 4 |
| chr12_+_51632666 | 0.10 |
ENST00000604900.1 |
DAZAP2 |
DAZ associated protein 2 |
| chr4_+_38665810 | 0.10 |
ENST00000261438.5 ENST00000514033.1 |
KLF3 |
Kruppel-like factor 3 (basic) |
| chr11_-_44971702 | 0.10 |
ENST00000533940.1 ENST00000533937.1 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr1_-_156647189 | 0.10 |
ENST00000368223.3 |
NES |
nestin |
| chr4_-_42154895 | 0.09 |
ENST00000502486.1 ENST00000504360.1 |
BEND4 |
BEN domain containing 4 |
| chr1_-_200379129 | 0.09 |
ENST00000367353.1 |
ZNF281 |
zinc finger protein 281 |
| chr11_+_60691924 | 0.09 |
ENST00000544065.1 ENST00000453848.2 ENST00000005286.4 |
TMEM132A |
transmembrane protein 132A |
| chr14_-_107078851 | 0.09 |
ENST00000390628.2 |
IGHV1-58 |
immunoglobulin heavy variable 1-58 |
| chr21_-_43187231 | 0.09 |
ENST00000332512.3 ENST00000352483.2 |
RIPK4 |
receptor-interacting serine-threonine kinase 4 |
| chr1_-_109203648 | 0.09 |
ENST00000370031.1 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr1_-_161014731 | 0.09 |
ENST00000368020.1 |
USF1 |
upstream transcription factor 1 |
| chr1_-_200379180 | 0.08 |
ENST00000294740.3 |
ZNF281 |
zinc finger protein 281 |
| chr15_+_22736484 | 0.08 |
ENST00000560659.2 |
GOLGA6L1 |
golgin A6 family-like 1 |
| chrX_-_153151586 | 0.08 |
ENST00000370060.1 ENST00000370055.1 ENST00000420165.1 |
L1CAM |
L1 cell adhesion molecule |
| chr1_-_93426998 | 0.08 |
ENST00000370310.4 |
FAM69A |
family with sequence similarity 69, member A |
| chr17_-_42031300 | 0.08 |
ENST00000592796.1 |
PYY |
peptide YY |
| chr1_-_109203997 | 0.08 |
ENST00000370032.5 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr14_-_23398565 | 0.08 |
ENST00000397440.4 ENST00000538452.1 ENST00000421938.2 ENST00000554867.1 ENST00000556616.1 ENST00000216350.8 ENST00000553550.1 ENST00000397441.2 ENST00000553897.1 |
PRMT5 |
protein arginine methyltransferase 5 |
| chr16_-_11680791 | 0.08 |
ENST00000571976.1 ENST00000413364.2 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr7_-_81399411 | 0.08 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr12_+_122688090 | 0.08 |
ENST00000324189.4 ENST00000546192.1 |
B3GNT4 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr11_+_68228186 | 0.08 |
ENST00000393799.2 ENST00000393800.2 ENST00000528635.1 ENST00000533127.1 ENST00000529907.1 ENST00000529344.1 ENST00000534534.1 ENST00000524845.1 ENST00000265637.4 ENST00000524904.1 ENST00000393801.3 ENST00000265636.5 ENST00000529710.1 |
PPP6R3 |
protein phosphatase 6, regulatory subunit 3 |
| chr10_-_52645379 | 0.08 |
ENST00000395489.2 |
A1CF |
APOBEC1 complementation factor |
| chr3_+_40428647 | 0.07 |
ENST00000301825.3 ENST00000439533.1 ENST00000456402.1 |
ENTPD3 |
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr5_+_126626498 | 0.07 |
ENST00000503335.2 ENST00000508365.1 ENST00000418761.2 ENST00000274473.6 |
MEGF10 |
multiple EGF-like-domains 10 |
| chr1_+_45965725 | 0.07 |
ENST00000401061.4 |
MMACHC |
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr1_+_6508100 | 0.07 |
ENST00000461727.1 |
ESPN |
espin |
| chrX_-_102319092 | 0.07 |
ENST00000372728.3 |
BEX1 |
brain expressed, X-linked 1 |
| chr12_+_50017184 | 0.07 |
ENST00000548825.2 |
PRPF40B |
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr17_+_7348658 | 0.07 |
ENST00000570557.1 ENST00000536404.2 ENST00000576360.1 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr9_+_139780942 | 0.07 |
ENST00000247668.2 ENST00000359662.3 |
TRAF2 |
TNF receptor-associated factor 2 |
| chr3_+_49726932 | 0.07 |
ENST00000327697.6 ENST00000432042.1 ENST00000454491.1 |
RNF123 |
ring finger protein 123 |
| chr1_+_11796177 | 0.07 |
ENST00000400895.2 ENST00000376629.4 ENST00000376627.2 ENST00000314340.5 ENST00000452018.2 ENST00000510878.1 |
AGTRAP |
angiotensin II receptor-associated protein |
| chr1_-_113498943 | 0.07 |
ENST00000369626.3 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr9_+_125281420 | 0.07 |
ENST00000340750.1 |
OR1J4 |
olfactory receptor, family 1, subfamily J, member 4 |
| chr19_+_37342547 | 0.07 |
ENST00000331800.4 ENST00000586646.1 |
ZNF345 |
zinc finger protein 345 |
| chr1_-_222763101 | 0.07 |
ENST00000391883.2 ENST00000366890.1 |
TAF1A |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr6_-_29324054 | 0.07 |
ENST00000543825.1 |
OR5V1 |
olfactory receptor, family 5, subfamily V, member 1 |
| chr11_-_19082216 | 0.07 |
ENST00000329773.2 |
MRGPRX2 |
MAS-related GPR, member X2 |
| chr16_-_11680759 | 0.07 |
ENST00000571459.1 ENST00000570798.1 ENST00000572255.1 ENST00000574763.1 ENST00000574703.1 ENST00000571277.1 ENST00000381810.3 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr6_-_35888905 | 0.07 |
ENST00000510290.1 ENST00000423325.2 ENST00000373822.1 |
SRPK1 |
SRSF protein kinase 1 |
| chrX_-_21776281 | 0.06 |
ENST00000379494.3 |
SMPX |
small muscle protein, X-linked |
| chr12_-_125348329 | 0.06 |
ENST00000546215.1 ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1 |
scavenger receptor class B, member 1 |
| chr17_+_37784749 | 0.06 |
ENST00000394265.1 ENST00000394267.2 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr1_-_40367668 | 0.06 |
ENST00000397332.2 ENST00000429311.1 |
MYCL |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr1_-_222763214 | 0.06 |
ENST00000350027.4 |
TAF1A |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr1_-_109203685 | 0.06 |
ENST00000402983.1 ENST00000420055.1 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr7_+_35756092 | 0.06 |
ENST00000458087.3 |
AC018647.3 |
AC018647.3 |
| chr1_-_222763240 | 0.06 |
ENST00000352967.4 ENST00000391882.1 ENST00000543857.1 |
TAF1A |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr9_-_123691047 | 0.06 |
ENST00000373887.3 |
TRAF1 |
TNF receptor-associated factor 1 |
| chr12_+_57998400 | 0.06 |
ENST00000548804.1 ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3 |
deltex homolog 3 (Drosophila) |
| chr7_+_35756186 | 0.06 |
ENST00000430518.1 |
AC018647.3 |
AC018647.3 |
| chr12_+_57998595 | 0.06 |
ENST00000337737.3 ENST00000548198.1 ENST00000551632.1 |
DTX3 |
deltex homolog 3 (Drosophila) |
| chrX_-_8700171 | 0.06 |
ENST00000262648.3 |
KAL1 |
Kallmann syndrome 1 sequence |
| chr9_+_21802542 | 0.06 |
ENST00000380172.4 |
MTAP |
methylthioadenosine phosphorylase |
| chr14_-_21493649 | 0.06 |
ENST00000553442.1 ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2 |
NDRG family member 2 |
| chr1_+_29241027 | 0.06 |
ENST00000373797.1 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr18_-_812231 | 0.05 |
ENST00000314574.4 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr17_-_62207485 | 0.05 |
ENST00000433197.3 |
ERN1 |
endoplasmic reticulum to nucleus signaling 1 |
| chr11_+_33037401 | 0.05 |
ENST00000241051.3 |
DEPDC7 |
DEP domain containing 7 |
| chr6_+_111303218 | 0.05 |
ENST00000441448.2 |
RPF2 |
ribosome production factor 2 homolog (S. cerevisiae) |
| chr14_+_103566665 | 0.05 |
ENST00000559116.1 |
EXOC3L4 |
exocyst complex component 3-like 4 |
| chr4_-_87855851 | 0.05 |
ENST00000473559.1 |
C4orf36 |
chromosome 4 open reading frame 36 |
| chr11_-_130184470 | 0.05 |
ENST00000357899.4 ENST00000397753.1 |
ZBTB44 |
zinc finger and BTB domain containing 44 |
| chr1_+_160709055 | 0.05 |
ENST00000368043.3 ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7 |
SLAM family member 7 |
| chr3_-_122512619 | 0.05 |
ENST00000383659.1 ENST00000306103.2 |
HSPBAP1 |
HSPB (heat shock 27kDa) associated protein 1 |
| chr3_+_184032919 | 0.05 |
ENST00000427845.1 ENST00000342981.4 ENST00000319274.6 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr16_-_15188106 | 0.05 |
ENST00000429751.2 ENST00000564131.1 ENST00000563559.1 ENST00000198767.6 |
RRN3 |
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr1_+_165796753 | 0.05 |
ENST00000367879.4 |
UCK2 |
uridine-cytidine kinase 2 |
| chr9_+_139873264 | 0.05 |
ENST00000446677.1 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chrX_+_21392529 | 0.05 |
ENST00000425654.2 ENST00000543067.1 |
CNKSR2 |
connector enhancer of kinase suppressor of Ras 2 |
| chr2_-_40006357 | 0.05 |
ENST00000505747.1 |
THUMPD2 |
THUMP domain containing 2 |
| chr6_+_31021973 | 0.05 |
ENST00000570223.1 ENST00000566475.1 ENST00000426185.1 |
HCG22 |
HLA complex group 22 |
| chr10_-_101380121 | 0.05 |
ENST00000370495.4 |
SLC25A28 |
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr2_-_241080069 | 0.05 |
ENST00000319460.1 |
OTOS |
otospiralin |
| chr11_+_63137251 | 0.05 |
ENST00000310969.4 ENST00000279178.3 |
SLC22A9 |
solute carrier family 22 (organic anion transporter), member 9 |
| chr6_-_41040268 | 0.05 |
ENST00000373154.2 ENST00000244558.9 ENST00000464633.1 ENST00000424266.2 ENST00000479950.1 ENST00000482515.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chr4_-_2936514 | 0.05 |
ENST00000508221.1 ENST00000507555.1 ENST00000355443.4 |
MFSD10 |
major facilitator superfamily domain containing 10 |
| chr3_-_52486841 | 0.05 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr10_+_70715884 | 0.05 |
ENST00000354185.4 |
DDX21 |
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr17_-_12921270 | 0.05 |
ENST00000578071.1 ENST00000426905.3 ENST00000395962.2 ENST00000583371.1 ENST00000338034.4 |
ELAC2 |
elaC ribonuclease Z 2 |
| chrX_-_49042778 | 0.05 |
ENST00000538114.1 ENST00000376310.3 ENST00000376317.3 ENST00000417014.1 |
PRICKLE3 |
prickle homolog 3 (Drosophila) |
| chr6_+_96025341 | 0.05 |
ENST00000369293.1 ENST00000358812.4 |
MANEA |
mannosidase, endo-alpha |
| chrX_-_40506766 | 0.05 |
ENST00000378421.1 ENST00000440784.2 ENST00000327877.5 ENST00000378426.1 ENST00000378418.2 |
CXorf38 |
chromosome X open reading frame 38 |
| chr1_-_31902614 | 0.05 |
ENST00000596131.1 |
AC114494.1 |
HCG1787699; Uncharacterized protein |
| chr3_-_55521323 | 0.05 |
ENST00000264634.4 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
| chr6_+_19837592 | 0.05 |
ENST00000378700.3 |
ID4 |
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr3_-_118959733 | 0.05 |
ENST00000459778.1 ENST00000359213.3 |
B4GALT4 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr14_-_77787198 | 0.05 |
ENST00000261534.4 |
POMT2 |
protein-O-mannosyltransferase 2 |
| chr17_+_36584662 | 0.04 |
ENST00000431231.2 ENST00000437668.3 |
ARHGAP23 |
Rho GTPase activating protein 23 |
| chr18_-_812517 | 0.04 |
ENST00000584307.1 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr11_+_9482551 | 0.04 |
ENST00000438144.2 ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143 |
zinc finger protein 143 |
| chr7_+_36192855 | 0.04 |
ENST00000534978.1 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr1_-_113498616 | 0.04 |
ENST00000433570.4 ENST00000538576.1 ENST00000458229.1 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr16_-_3086927 | 0.04 |
ENST00000572449.1 |
CCDC64B |
coiled-coil domain containing 64B |
| chr11_+_64073699 | 0.04 |
ENST00000405666.1 ENST00000468670.1 |
ESRRA |
estrogen-related receptor alpha |
| chr22_-_29138386 | 0.04 |
ENST00000544772.1 |
CHEK2 |
checkpoint kinase 2 |
| chr6_-_31324943 | 0.04 |
ENST00000412585.2 ENST00000434333.1 |
HLA-B |
major histocompatibility complex, class I, B |
| chr16_-_11836595 | 0.04 |
ENST00000356957.3 ENST00000283033.5 |
TXNDC11 |
thioredoxin domain containing 11 |
| chr16_+_69985083 | 0.04 |
ENST00000288040.6 ENST00000449317.2 |
CLEC18A |
C-type lectin domain family 18, member A |
| chr22_+_29279552 | 0.04 |
ENST00000544604.2 |
ZNRF3 |
zinc and ring finger 3 |
| chr11_-_58980342 | 0.04 |
ENST00000361050.3 |
MPEG1 |
macrophage expressed 1 |
| chr19_-_7553852 | 0.04 |
ENST00000593547.1 |
PEX11G |
peroxisomal biogenesis factor 11 gamma |
| chr3_+_118905564 | 0.04 |
ENST00000460625.1 |
UPK1B |
uroplakin 1B |
| chr7_+_148395733 | 0.04 |
ENST00000602748.1 |
CUL1 |
cullin 1 |
| chr19_+_49661037 | 0.04 |
ENST00000427978.2 |
TRPM4 |
transient receptor potential cation channel, subfamily M, member 4 |
| chr6_+_41040678 | 0.04 |
ENST00000341376.6 ENST00000353205.5 |
NFYA |
nuclear transcription factor Y, alpha |
| chr18_-_19997878 | 0.04 |
ENST00000391403.2 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr22_+_26879817 | 0.04 |
ENST00000215917.7 |
SRRD |
SRR1 domain containing |
| chr14_-_24740709 | 0.04 |
ENST00000399409.3 ENST00000216840.6 |
RABGGTA |
Rab geranylgeranyltransferase, alpha subunit |
| chr5_+_43192311 | 0.04 |
ENST00000326035.2 |
NIM1 |
NIM1 serine/threonine protein kinase |
| chr8_+_87354945 | 0.04 |
ENST00000517970.1 |
WWP1 |
WW domain containing E3 ubiquitin protein ligase 1 |
| chr10_-_52645416 | 0.04 |
ENST00000374001.2 ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF |
APOBEC1 complementation factor |
| chr12_-_133613794 | 0.04 |
ENST00000443154.3 |
RP11-386I8.6 |
RP11-386I8.6 |
| chr9_+_133305669 | 0.04 |
ENST00000302481.1 |
HMCN2 |
hemicentin 2 |
| chr7_+_148395959 | 0.04 |
ENST00000325222.4 |
CUL1 |
cullin 1 |
| chr12_+_50478977 | 0.04 |
ENST00000381513.4 |
SMARCD1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr7_+_156742399 | 0.04 |
ENST00000275820.3 |
NOM1 |
nucleolar protein with MIF4G domain 1 |
| chr1_-_40367530 | 0.04 |
ENST00000372816.2 ENST00000372815.1 |
MYCL |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr12_+_57916466 | 0.04 |
ENST00000355673.3 |
MBD6 |
methyl-CpG binding domain protein 6 |
| chr12_+_125478241 | 0.04 |
ENST00000341446.8 |
BRI3BP |
BRI3 binding protein |
| chr7_-_105752971 | 0.04 |
ENST00000011473.2 |
SYPL1 |
synaptophysin-like 1 |
| chr10_+_120967072 | 0.04 |
ENST00000392870.2 |
GRK5 |
G protein-coupled receptor kinase 5 |
| chr1_-_59165763 | 0.04 |
ENST00000472487.1 |
MYSM1 |
Myb-like, SWIRM and MPN domains 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:2000412 | activation of MAPK activity involved in innate immune response(GO:0035419) positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) positive regulation of thymocyte migration(GO:2000412) |
| 0.2 | 0.3 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.4 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.3 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.1 | 0.3 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.2 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.4 | GO:0015820 | leucine transport(GO:0015820) |
| 0.0 | 0.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.0 | 0.2 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.7 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.0 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.1 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0046495 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.0 | 0.1 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.0 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.1 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.0 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.2 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.0 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.0 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.0 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:0046502 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0071065 | dense core granule membrane(GO:0032127) alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.0 | 0.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.2 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.2 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |