Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for REL
Z-value: 0.94
Transcription factors associated with REL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
REL
|
ENSG00000162924.9 | REL |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| REL | hg19_v2_chr2_+_61108650_61108687 | 0.97 | 9.9e-05 | Click! |
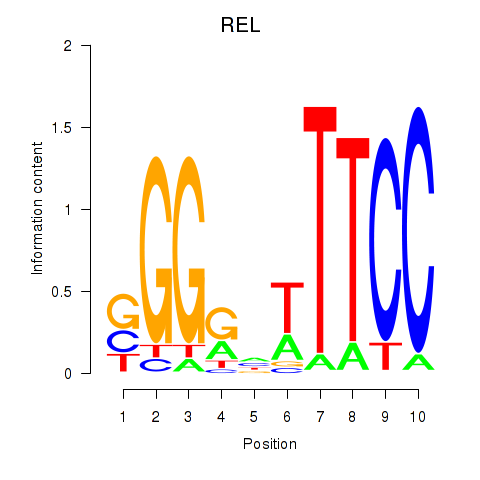
Activity profile of REL motif
Sorted Z-values of REL motif
Network of associatons between targets according to the STRING database.
First level regulatory network of REL
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_21529811 | 1.66 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr18_+_21452964 | 1.64 |
ENST00000587184.1 |
LAMA3 |
laminin, alpha 3 |
| chr18_+_21452804 | 1.47 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr6_+_138188551 | 1.22 |
ENST00000237289.4 ENST00000433680.1 |
TNFAIP3 |
tumor necrosis factor, alpha-induced protein 3 |
| chr2_-_113594279 | 0.83 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr10_-_100027943 | 0.79 |
ENST00000260702.3 |
LOXL4 |
lysyl oxidase-like 4 |
| chr18_+_33877654 | 0.75 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr20_+_43803517 | 0.61 |
ENST00000243924.3 |
PI3 |
peptidase inhibitor 3, skin-derived |
| chr1_-_209824643 | 0.60 |
ENST00000391911.1 ENST00000415782.1 |
LAMB3 |
laminin, beta 3 |
| chr4_+_74735102 | 0.60 |
ENST00000395761.3 |
CXCL1 |
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr7_+_69064300 | 0.58 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr17_-_56492989 | 0.56 |
ENST00000583753.1 |
RNF43 |
ring finger protein 43 |
| chr11_+_10476851 | 0.54 |
ENST00000396553.2 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr11_-_58345569 | 0.50 |
ENST00000528954.1 ENST00000528489.1 |
LPXN |
leupaxin |
| chr6_+_30850697 | 0.48 |
ENST00000509639.1 ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr12_-_95009837 | 0.48 |
ENST00000551457.1 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
| chr6_-_143832820 | 0.48 |
ENST00000002165.6 |
FUCA2 |
fucosidase, alpha-L- 2, plasma |
| chr2_+_163175394 | 0.43 |
ENST00000446271.1 ENST00000429691.2 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr1_+_95582881 | 0.42 |
ENST00000370203.4 ENST00000456991.1 |
TMEM56 |
transmembrane protein 56 |
| chr12_-_86230315 | 0.38 |
ENST00000361228.3 |
RASSF9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr12_+_51632666 | 0.37 |
ENST00000604900.1 |
DAZAP2 |
DAZ associated protein 2 |
| chr19_-_51471381 | 0.37 |
ENST00000594641.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr19_-_51471362 | 0.36 |
ENST00000376853.4 ENST00000424910.2 |
KLK6 |
kallikrein-related peptidase 6 |
| chr19_-_51472031 | 0.35 |
ENST00000391808.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr17_-_4643114 | 0.32 |
ENST00000293778.6 |
CXCL16 |
chemokine (C-X-C motif) ligand 16 |
| chr2_+_220094479 | 0.32 |
ENST00000323348.5 ENST00000453432.1 ENST00000409849.1 ENST00000416565.1 ENST00000410034.3 ENST00000447157.1 |
ANKZF1 |
ankyrin repeat and zinc finger domain containing 1 |
| chr11_+_102188224 | 0.32 |
ENST00000263464.3 |
BIRC3 |
baculoviral IAP repeat containing 3 |
| chr9_-_117880477 | 0.31 |
ENST00000534839.1 ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC |
tenascin C |
| chr9_+_130911770 | 0.31 |
ENST00000372998.1 |
LCN2 |
lipocalin 2 |
| chr1_-_204380919 | 0.30 |
ENST00000367188.4 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
| chr14_+_23842018 | 0.29 |
ENST00000397242.2 ENST00000329715.2 |
IL25 |
interleukin 25 |
| chr11_-_72492878 | 0.29 |
ENST00000535054.1 ENST00000545082.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr16_-_88717482 | 0.29 |
ENST00000261623.3 |
CYBA |
cytochrome b-245, alpha polypeptide |
| chr19_-_4338838 | 0.28 |
ENST00000594605.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chr8_+_123793633 | 0.28 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr12_-_122985067 | 0.28 |
ENST00000540586.1 ENST00000543897.1 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chr19_-_4338783 | 0.28 |
ENST00000601482.1 ENST00000600324.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chr12_-_55375622 | 0.28 |
ENST00000316577.8 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
| chr8_-_105601134 | 0.28 |
ENST00000276654.5 ENST00000424843.2 |
LRP12 |
low density lipoprotein receptor-related protein 12 |
| chr16_+_27325202 | 0.27 |
ENST00000395762.2 ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R |
interleukin 4 receptor |
| chr10_-_96122682 | 0.26 |
ENST00000371361.3 |
NOC3L |
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr9_-_136004782 | 0.26 |
ENST00000393157.3 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
| chr10_+_104155450 | 0.25 |
ENST00000471698.1 ENST00000189444.6 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr9_+_130911723 | 0.25 |
ENST00000277480.2 ENST00000373013.2 ENST00000540948.1 |
LCN2 |
lipocalin 2 |
| chr11_-_72492903 | 0.25 |
ENST00000537947.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr16_+_3014217 | 0.25 |
ENST00000572045.1 |
KREMEN2 |
kringle containing transmembrane protein 2 |
| chr3_+_183967409 | 0.25 |
ENST00000324557.4 ENST00000402825.3 |
ECE2 |
endothelin converting enzyme 2 |
| chr2_-_74669009 | 0.25 |
ENST00000272430.5 |
RTKN |
rhotekin |
| chr2_-_163175133 | 0.24 |
ENST00000421365.2 ENST00000263642.2 |
IFIH1 |
interferon induced with helicase C domain 1 |
| chr11_+_102188272 | 0.23 |
ENST00000532808.1 |
BIRC3 |
baculoviral IAP repeat containing 3 |
| chr6_+_15401075 | 0.23 |
ENST00000541660.1 |
JARID2 |
jumonji, AT rich interactive domain 2 |
| chr6_+_32121908 | 0.23 |
ENST00000375143.2 ENST00000424499.1 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr6_-_44233361 | 0.23 |
ENST00000275015.5 |
NFKBIE |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr6_+_32121789 | 0.22 |
ENST00000437001.2 ENST00000375137.2 |
PPT2 |
palmitoyl-protein thioesterase 2 |
| chr2_+_61108650 | 0.22 |
ENST00000295025.8 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr7_+_128784712 | 0.22 |
ENST00000289407.4 |
TSPAN33 |
tetraspanin 33 |
| chr14_+_103243813 | 0.22 |
ENST00000560371.1 ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3 |
TNF receptor-associated factor 3 |
| chr14_+_68086515 | 0.22 |
ENST00000261783.3 |
ARG2 |
arginase 2 |
| chr7_-_16844611 | 0.21 |
ENST00000401412.1 ENST00000419304.2 |
AGR2 |
anterior gradient 2 |
| chr1_-_93426998 | 0.21 |
ENST00000370310.4 |
FAM69A |
family with sequence similarity 69, member A |
| chr19_-_46272106 | 0.21 |
ENST00000560168.1 |
SIX5 |
SIX homeobox 5 |
| chr9_+_6328338 | 0.21 |
ENST00000344545.5 |
TPD52L3 |
tumor protein D52-like 3 |
| chr12_-_122985494 | 0.20 |
ENST00000336229.4 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chr21_+_46875424 | 0.20 |
ENST00000359759.4 |
COL18A1 |
collagen, type XVIII, alpha 1 |
| chr16_+_3096638 | 0.20 |
ENST00000336577.4 |
MMP25 |
matrix metallopeptidase 25 |
| chr19_+_14142535 | 0.20 |
ENST00000263379.2 |
IL27RA |
interleukin 27 receptor, alpha |
| chr3_-_183967296 | 0.20 |
ENST00000455059.1 ENST00000445626.2 |
ALG3 |
ALG3, alpha-1,3- mannosyltransferase |
| chr7_-_19184929 | 0.20 |
ENST00000275461.3 |
FERD3L |
Fer3-like bHLH transcription factor |
| chr6_-_29527702 | 0.19 |
ENST00000377050.4 |
UBD |
ubiquitin D |
| chr11_+_7597639 | 0.19 |
ENST00000533792.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_+_50497784 | 0.19 |
ENST00000548814.1 |
GPD1 |
glycerol-3-phosphate dehydrogenase 1 (soluble) |
| chr1_-_155959853 | 0.19 |
ENST00000462460.2 ENST00000368316.1 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr2_+_90077680 | 0.18 |
ENST00000390270.2 |
IGKV3D-20 |
immunoglobulin kappa variable 3D-20 |
| chr9_-_124990680 | 0.18 |
ENST00000541397.2 ENST00000560485.1 |
LHX6 |
LIM homeobox 6 |
| chr6_-_31550192 | 0.18 |
ENST00000429299.2 ENST00000446745.2 |
LTB |
lymphotoxin beta (TNF superfamily, member 3) |
| chr19_+_41222998 | 0.18 |
ENST00000263370.2 |
ITPKC |
inositol-trisphosphate 3-kinase C |
| chr16_-_75498553 | 0.18 |
ENST00000569276.1 ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A RP11-77K12.1 |
transmembrane protein 170A Uncharacterized protein |
| chr4_-_74864386 | 0.17 |
ENST00000296027.4 |
CXCL5 |
chemokine (C-X-C motif) ligand 5 |
| chr2_-_89247338 | 0.17 |
ENST00000496168.1 |
IGKV1-5 |
immunoglobulin kappa variable 1-5 |
| chr16_-_88717423 | 0.17 |
ENST00000568278.1 ENST00000569359.1 ENST00000567174.1 |
CYBA |
cytochrome b-245, alpha polypeptide |
| chr14_+_103589789 | 0.17 |
ENST00000558056.1 ENST00000560869.1 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
| chrX_-_40594755 | 0.17 |
ENST00000324817.1 |
MED14 |
mediator complex subunit 14 |
| chr15_+_42131011 | 0.17 |
ENST00000458483.1 |
PLA2G4B |
phospholipase A2, group IVB (cytosolic) |
| chr6_+_135502408 | 0.17 |
ENST00000341911.5 ENST00000442647.2 ENST00000316528.8 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr6_+_151042224 | 0.17 |
ENST00000358517.2 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr2_+_102314161 | 0.16 |
ENST00000425019.1 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr10_+_104154229 | 0.16 |
ENST00000428099.1 ENST00000369966.3 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr20_+_42984330 | 0.16 |
ENST00000316673.4 ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr12_-_117537240 | 0.15 |
ENST00000392545.4 ENST00000541210.1 ENST00000335209.7 |
TESC |
tescalcin |
| chr7_-_126892303 | 0.15 |
ENST00000358373.3 |
GRM8 |
glutamate receptor, metabotropic 8 |
| chr14_-_24658053 | 0.15 |
ENST00000354464.6 |
IPO4 |
importin 4 |
| chr4_-_76957214 | 0.15 |
ENST00000306621.3 |
CXCL11 |
chemokine (C-X-C motif) ligand 11 |
| chr12_+_56211703 | 0.15 |
ENST00000243045.5 ENST00000552672.1 ENST00000550836.1 |
ORMDL2 |
ORM1-like 2 (S. cerevisiae) |
| chr14_-_69446034 | 0.15 |
ENST00000193403.6 |
ACTN1 |
actinin, alpha 1 |
| chr7_-_98741714 | 0.15 |
ENST00000361125.1 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
| chr9_-_34637806 | 0.15 |
ENST00000477726.1 |
SIGMAR1 |
sigma non-opioid intracellular receptor 1 |
| chr21_-_34915147 | 0.14 |
ENST00000381831.3 ENST00000381839.3 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr1_+_156117149 | 0.14 |
ENST00000435124.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr2_-_220094031 | 0.14 |
ENST00000443140.1 ENST00000432520.1 ENST00000409618.1 |
ATG9A |
autophagy related 9A |
| chr22_+_46731596 | 0.14 |
ENST00000381019.3 |
TRMU |
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr2_-_220094294 | 0.13 |
ENST00000436856.1 ENST00000428226.1 ENST00000409422.1 ENST00000431715.1 ENST00000457841.1 ENST00000439812.1 ENST00000361242.4 ENST00000396761.2 |
ATG9A |
autophagy related 9A |
| chr1_+_144339738 | 0.13 |
ENST00000538264.1 |
AL592284.1 |
Protein LOC642441 |
| chr12_+_7055631 | 0.13 |
ENST00000543115.1 ENST00000399448.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr14_+_77648167 | 0.13 |
ENST00000554346.1 ENST00000298351.4 |
TMEM63C |
transmembrane protein 63C |
| chr7_-_98741642 | 0.13 |
ENST00000361368.2 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
| chr1_-_89738528 | 0.12 |
ENST00000343435.5 |
GBP5 |
guanylate binding protein 5 |
| chr16_-_2059748 | 0.12 |
ENST00000562103.1 ENST00000431526.1 |
ZNF598 |
zinc finger protein 598 |
| chr6_-_32821599 | 0.12 |
ENST00000354258.4 |
TAP1 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_-_104893863 | 0.12 |
ENST00000260315.3 ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5 |
caspase 5, apoptosis-related cysteine peptidase |
| chr4_-_74904398 | 0.12 |
ENST00000296026.4 |
CXCL3 |
chemokine (C-X-C motif) ligand 3 |
| chr3_-_49142178 | 0.12 |
ENST00000452739.1 ENST00000414533.1 ENST00000417025.1 |
QARS |
glutaminyl-tRNA synthetase |
| chr3_-_49142504 | 0.11 |
ENST00000306125.6 ENST00000420147.2 |
QARS |
glutaminyl-tRNA synthetase |
| chr2_+_90153696 | 0.11 |
ENST00000417279.2 |
IGKV3D-15 |
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr12_-_7281469 | 0.11 |
ENST00000542370.1 ENST00000266560.3 |
RBP5 |
retinol binding protein 5, cellular |
| chr1_+_179050512 | 0.11 |
ENST00000367627.3 |
TOR3A |
torsin family 3, member A |
| chr5_-_149792295 | 0.11 |
ENST00000518797.1 ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr20_-_43977055 | 0.11 |
ENST00000372733.3 ENST00000537976.1 |
SDC4 |
syndecan 4 |
| chr9_-_115095883 | 0.11 |
ENST00000450374.1 ENST00000374255.2 ENST00000334318.6 ENST00000374257.1 |
PTBP3 |
polypyrimidine tract binding protein 3 |
| chr16_+_84209539 | 0.11 |
ENST00000569735.1 |
DNAAF1 |
dynein, axonemal, assembly factor 1 |
| chr12_-_62997214 | 0.11 |
ENST00000408887.2 |
C12orf61 |
chromosome 12 open reading frame 61 |
| chr22_-_38380543 | 0.11 |
ENST00000396884.2 |
SOX10 |
SRY (sex determining region Y)-box 10 |
| chr10_-_108924284 | 0.10 |
ENST00000344440.6 ENST00000263054.6 |
SORCS1 |
sortilin-related VPS10 domain containing receptor 1 |
| chr3_-_119379719 | 0.10 |
ENST00000493094.1 |
POPDC2 |
popeye domain containing 2 |
| chr19_-_49250054 | 0.10 |
ENST00000602105.1 ENST00000332955.2 |
IZUMO1 |
izumo sperm-egg fusion 1 |
| chr20_-_36156125 | 0.10 |
ENST00000397135.1 ENST00000397137.1 |
BLCAP |
bladder cancer associated protein |
| chr12_+_53491220 | 0.10 |
ENST00000548547.1 ENST00000301464.3 |
IGFBP6 |
insulin-like growth factor binding protein 6 |
| chr19_+_39390587 | 0.10 |
ENST00000572515.1 ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr7_+_156742399 | 0.10 |
ENST00000275820.3 |
NOM1 |
nucleolar protein with MIF4G domain 1 |
| chr17_+_49337881 | 0.10 |
ENST00000225298.7 |
UTP18 |
UTP18 small subunit (SSU) processome component homolog (yeast) |
| chr13_-_52027134 | 0.10 |
ENST00000311234.4 ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6 |
integrator complex subunit 6 |
| chr5_-_35230434 | 0.10 |
ENST00000504500.1 |
PRLR |
prolactin receptor |
| chr19_-_4831701 | 0.09 |
ENST00000248244.5 |
TICAM1 |
toll-like receptor adaptor molecule 1 |
| chr2_-_113993020 | 0.09 |
ENST00000465084.1 |
PAX8 |
paired box 8 |
| chrX_-_17878827 | 0.09 |
ENST00000360011.1 |
RAI2 |
retinoic acid induced 2 |
| chr2_-_68694390 | 0.09 |
ENST00000377957.3 |
FBXO48 |
F-box protein 48 |
| chr19_+_4229495 | 0.09 |
ENST00000221847.5 |
EBI3 |
Epstein-Barr virus induced 3 |
| chr2_-_89310012 | 0.09 |
ENST00000493819.1 |
IGKV1-9 |
immunoglobulin kappa variable 1-9 |
| chr19_+_41256764 | 0.09 |
ENST00000243563.3 ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
| chr11_+_119039414 | 0.09 |
ENST00000409991.1 ENST00000292199.2 ENST00000409265.4 ENST00000409109.1 |
NLRX1 |
NLR family member X1 |
| chr20_-_36156293 | 0.09 |
ENST00000373537.2 ENST00000414542.2 |
BLCAP |
bladder cancer associated protein |
| chr16_+_84209738 | 0.09 |
ENST00000564928.1 |
DNAAF1 |
dynein, axonemal, assembly factor 1 |
| chr1_+_63833261 | 0.09 |
ENST00000371108.4 |
ALG6 |
ALG6, alpha-1,3-glucosyltransferase |
| chr3_+_101280677 | 0.09 |
ENST00000309922.6 ENST00000495642.1 |
TRMT10C |
tRNA methyltransferase 10 homolog C (S. cerevisiae) |
| chr2_+_16080659 | 0.09 |
ENST00000281043.3 |
MYCN |
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr14_+_37667230 | 0.09 |
ENST00000556451.1 ENST00000556753.1 ENST00000396294.2 |
MIPOL1 |
mirror-image polydactyly 1 |
| chrX_+_115567767 | 0.08 |
ENST00000371900.4 |
SLC6A14 |
solute carrier family 6 (amino acid transporter), member 14 |
| chr14_-_35873856 | 0.08 |
ENST00000553342.1 ENST00000216797.5 ENST00000557140.1 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr12_-_76425368 | 0.08 |
ENST00000602540.1 |
PHLDA1 |
pleckstrin homology-like domain, family A, member 1 |
| chr7_+_107384579 | 0.08 |
ENST00000222597.2 ENST00000415884.2 |
CBLL1 |
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr9_-_115095123 | 0.08 |
ENST00000458258.1 |
PTBP3 |
polypyrimidine tract binding protein 3 |
| chr21_+_34638656 | 0.08 |
ENST00000290200.2 |
IL10RB |
interleukin 10 receptor, beta |
| chr10_+_70587279 | 0.08 |
ENST00000298596.6 ENST00000399169.4 ENST00000399165.4 ENST00000399162.2 |
STOX1 |
storkhead box 1 |
| chr9_-_136344197 | 0.08 |
ENST00000414172.1 ENST00000371897.4 |
SLC2A6 |
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr2_+_173940668 | 0.08 |
ENST00000375213.3 |
MLTK |
Mitogen-activated protein kinase kinase kinase MLT |
| chr16_+_67571351 | 0.08 |
ENST00000428437.2 ENST00000569253.1 |
FAM65A |
family with sequence similarity 65, member A |
| chr4_+_74702214 | 0.08 |
ENST00000226317.5 ENST00000515050.1 |
CXCL6 |
chemokine (C-X-C motif) ligand 6 |
| chr4_-_185395672 | 0.08 |
ENST00000393593.3 |
IRF2 |
interferon regulatory factor 2 |
| chr16_+_50730910 | 0.07 |
ENST00000300589.2 |
NOD2 |
nucleotide-binding oligomerization domain containing 2 |
| chr2_+_114647504 | 0.07 |
ENST00000263238.2 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
| chr22_+_22676808 | 0.07 |
ENST00000390290.2 |
IGLV1-51 |
immunoglobulin lambda variable 1-51 |
| chr2_+_238600933 | 0.07 |
ENST00000420665.1 ENST00000392000.4 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
| chr5_+_49963239 | 0.07 |
ENST00000505554.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr15_-_73661605 | 0.07 |
ENST00000261917.3 |
HCN4 |
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr11_-_66313699 | 0.07 |
ENST00000526986.1 ENST00000310442.3 |
ZDHHC24 |
zinc finger, DHHC-type containing 24 |
| chr19_+_58193337 | 0.07 |
ENST00000601064.1 ENST00000282296.5 ENST00000356715.4 |
ZNF551 |
zinc finger protein 551 |
| chr1_-_62190793 | 0.07 |
ENST00000371177.2 ENST00000606498.1 |
TM2D1 |
TM2 domain containing 1 |
| chr1_-_6453426 | 0.07 |
ENST00000545482.1 |
ACOT7 |
acyl-CoA thioesterase 7 |
| chr6_+_29910301 | 0.07 |
ENST00000376809.5 ENST00000376802.2 |
HLA-A |
major histocompatibility complex, class I, A |
| chr15_+_52311398 | 0.07 |
ENST00000261845.5 |
MAPK6 |
mitogen-activated protein kinase 6 |
| chr12_-_40013553 | 0.07 |
ENST00000308666.3 |
ABCD2 |
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr16_+_88636789 | 0.07 |
ENST00000301011.5 ENST00000452588.2 |
ZC3H18 |
zinc finger CCCH-type containing 18 |
| chr10_-_18948156 | 0.07 |
ENST00000414939.1 ENST00000449529.1 ENST00000456217.1 ENST00000444660.1 |
ARL5B-AS1 |
ARL5B antisense RNA 1 |
| chr2_-_29093132 | 0.07 |
ENST00000306108.5 |
TRMT61B |
tRNA methyltransferase 61 homolog B (S. cerevisiae) |
| chr9_+_130547958 | 0.06 |
ENST00000421939.1 ENST00000373265.2 |
CDK9 |
cyclin-dependent kinase 9 |
| chr2_+_202047843 | 0.06 |
ENST00000272879.5 ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr19_+_35842445 | 0.06 |
ENST00000246553.2 |
FFAR1 |
free fatty acid receptor 1 |
| chr6_-_29399744 | 0.06 |
ENST00000377154.1 |
OR5V1 |
olfactory receptor, family 5, subfamily V, member 1 |
| chr17_-_6338399 | 0.06 |
ENST00000570584.1 ENST00000574913.1 ENST00000571740.1 ENST00000575265.1 ENST00000574506.1 |
AIPL1 |
aryl hydrocarbon receptor interacting protein-like 1 |
| chr2_-_177502659 | 0.06 |
ENST00000295549.4 |
AC017048.3 |
long intergenic non-protein coding RNA 1116 |
| chr2_+_202047596 | 0.06 |
ENST00000286186.6 ENST00000360132.3 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr1_+_201979645 | 0.06 |
ENST00000367284.5 ENST00000367283.3 |
ELF3 |
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr13_+_97928395 | 0.06 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr9_-_115095851 | 0.06 |
ENST00000343327.2 |
PTBP3 |
polypyrimidine tract binding protein 3 |
| chr4_+_72897521 | 0.05 |
ENST00000308744.6 ENST00000344413.5 |
NPFFR2 |
neuropeptide FF receptor 2 |
| chr1_+_165796753 | 0.05 |
ENST00000367879.4 |
UCK2 |
uridine-cytidine kinase 2 |
| chr5_+_150591678 | 0.05 |
ENST00000523466.1 |
GM2A |
GM2 ganglioside activator |
| chr18_-_31802282 | 0.05 |
ENST00000535475.1 |
NOL4 |
nucleolar protein 4 |
| chr2_+_63277927 | 0.05 |
ENST00000282549.2 |
OTX1 |
orthodenticle homeobox 1 |
| chr9_-_139927462 | 0.05 |
ENST00000314412.6 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
| chr3_+_93698974 | 0.05 |
ENST00000535334.1 ENST00000478400.1 ENST00000303097.7 ENST00000394222.3 ENST00000471138.1 ENST00000539730.1 |
ARL13B |
ADP-ribosylation factor-like 13B |
| chr17_+_80477571 | 0.05 |
ENST00000335255.5 |
FOXK2 |
forkhead box K2 |
| chr14_+_37667118 | 0.05 |
ENST00000556615.1 ENST00000327441.7 ENST00000536774.1 |
MIPOL1 |
mirror-image polydactyly 1 |
| chr18_-_31802056 | 0.05 |
ENST00000538587.1 |
NOL4 |
nucleolar protein 4 |
| chr1_-_235813290 | 0.05 |
ENST00000391854.2 |
GNG4 |
guanine nucleotide binding protein (G protein), gamma 4 |
| chr15_-_51058005 | 0.05 |
ENST00000261854.5 |
SPPL2A |
signal peptide peptidase like 2A |
| chr1_+_202385953 | 0.05 |
ENST00000466968.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr3_+_9932238 | 0.05 |
ENST00000307768.4 |
JAGN1 |
jagunal homolog 1 (Drosophila) |
| chr3_-_139108475 | 0.04 |
ENST00000515006.1 ENST00000513274.1 ENST00000514508.1 ENST00000507777.1 ENST00000512153.1 ENST00000333188.5 |
COPB2 |
coatomer protein complex, subunit beta 2 (beta prime) |
| chr19_+_35940486 | 0.04 |
ENST00000246549.2 |
FFAR2 |
free fatty acid receptor 2 |
| chr22_-_41252962 | 0.04 |
ENST00000216218.3 |
ST13 |
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0071947 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.3 | 0.8 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.2 | 0.6 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.2 | 5.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.6 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.5 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.1 | 0.3 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.3 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.5 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.1 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.1 | 0.6 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 0.2 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.3 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.3 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.2 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.5 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.1 | 0.2 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.2 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.3 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 1.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.5 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.5 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.5 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0009624 | response to nematode(GO:0009624) eosinophil differentiation(GO:0030222) |
| 0.0 | 0.2 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.2 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.2 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 1.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.3 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.2 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.2 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.6 | GO:0007620 | copulation(GO:0007620) |
| 0.0 | 0.2 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.2 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.0 | 0.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.3 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.2 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.1 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.0 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0002829 | negative regulation of type 2 immune response(GO:0002829) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:1901858 | cellular response to nitrosative stress(GO:0071500) regulation of mitochondrial DNA metabolic process(GO:1901858) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.3 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 1.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 1.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.8 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.2 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 5.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.6 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |