Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
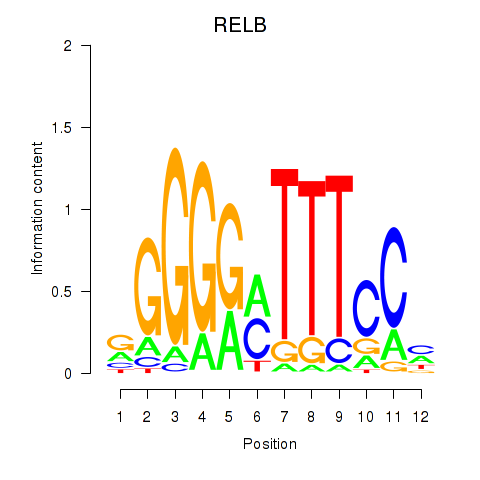
Results for RELB
Z-value: 0.15
Transcription factors associated with RELB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RELB
|
ENSG00000104856.9 | RELB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RELB | hg19_v2_chr19_+_45504688_45504782 | 0.48 | 2.3e-01 | Click! |
Activity profile of RELB motif
Sorted Z-values of RELB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RELB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_132891461 | 0.59 |
ENST00000275198.1 |
TAAR6 |
trace amine associated receptor 6 |
| chr8_-_57472154 | 0.28 |
ENST00000499425.1 ENST00000523664.1 ENST00000518943.1 ENST00000524338.1 |
LINC00968 |
long intergenic non-protein coding RNA 968 |
| chr1_+_6845578 | 0.25 |
ENST00000467404.2 ENST00000439411.2 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr9_+_127539481 | 0.25 |
ENST00000373580.3 |
OLFML2A |
olfactomedin-like 2A |
| chr1_-_113253977 | 0.23 |
ENST00000606505.1 ENST00000605933.1 |
RP11-426L16.10 |
Rho-related GTP-binding protein RhoC |
| chr1_+_6845497 | 0.22 |
ENST00000473578.1 ENST00000557126.1 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr6_+_32821924 | 0.21 |
ENST00000374859.2 ENST00000453265.2 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chrX_+_103031421 | 0.20 |
ENST00000433491.1 ENST00000418604.1 ENST00000443502.1 |
PLP1 |
proteolipid protein 1 |
| chr5_-_35230434 | 0.20 |
ENST00000504500.1 |
PRLR |
prolactin receptor |
| chr1_-_6321035 | 0.18 |
ENST00000377893.2 |
GPR153 |
G protein-coupled receptor 153 |
| chr2_-_56150184 | 0.16 |
ENST00000394554.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr12_-_8815215 | 0.16 |
ENST00000544889.1 ENST00000543369.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr12_-_8815299 | 0.16 |
ENST00000535336.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr6_-_24358264 | 0.16 |
ENST00000378454.3 |
DCDC2 |
doublecortin domain containing 2 |
| chr14_-_35873856 | 0.16 |
ENST00000553342.1 ENST00000216797.5 ENST00000557140.1 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr2_+_68870721 | 0.16 |
ENST00000303786.3 |
PROKR1 |
prokineticin receptor 1 |
| chr10_-_102089729 | 0.15 |
ENST00000465680.2 |
PKD2L1 |
polycystic kidney disease 2-like 1 |
| chr2_-_163175133 | 0.15 |
ENST00000421365.2 ENST00000263642.2 |
IFIH1 |
interferon induced with helicase C domain 1 |
| chr10_+_12391685 | 0.15 |
ENST00000378845.1 |
CAMK1D |
calcium/calmodulin-dependent protein kinase ID |
| chr9_+_101705893 | 0.15 |
ENST00000375001.3 |
COL15A1 |
collagen, type XV, alpha 1 |
| chr3_+_52280173 | 0.14 |
ENST00000296487.4 |
PPM1M |
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr1_+_6845384 | 0.14 |
ENST00000303635.7 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr3_+_52280220 | 0.13 |
ENST00000409502.3 ENST00000323588.4 |
PPM1M |
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr21_+_46875424 | 0.13 |
ENST00000359759.4 |
COL18A1 |
collagen, type XVIII, alpha 1 |
| chr21_+_46875395 | 0.13 |
ENST00000355480.5 |
COL18A1 |
collagen, type XVIII, alpha 1 |
| chr19_-_46272462 | 0.13 |
ENST00000317578.6 |
SIX5 |
SIX homeobox 5 |
| chr19_-_9879293 | 0.12 |
ENST00000397902.2 ENST00000592859.1 ENST00000588267.1 |
ZNF846 |
zinc finger protein 846 |
| chr19_+_41222998 | 0.12 |
ENST00000263370.2 |
ITPKC |
inositol-trisphosphate 3-kinase C |
| chr11_+_18287721 | 0.12 |
ENST00000356524.4 |
SAA1 |
serum amyloid A1 |
| chr11_+_706219 | 0.12 |
ENST00000533500.1 |
EPS8L2 |
EPS8-like 2 |
| chr16_-_4850471 | 0.12 |
ENST00000592019.1 ENST00000586153.1 |
ROGDI |
rogdi homolog (Drosophila) |
| chr11_+_18287801 | 0.12 |
ENST00000532858.1 ENST00000405158.2 |
SAA1 |
serum amyloid A1 |
| chr1_-_209979375 | 0.12 |
ENST00000367021.3 |
IRF6 |
interferon regulatory factor 6 |
| chr10_+_12391481 | 0.12 |
ENST00000378847.3 |
CAMK1D |
calcium/calmodulin-dependent protein kinase ID |
| chr2_+_163175394 | 0.12 |
ENST00000446271.1 ENST00000429691.2 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr7_+_69064300 | 0.12 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr8_-_9008206 | 0.11 |
ENST00000310455.3 |
PPP1R3B |
protein phosphatase 1, regulatory subunit 3B |
| chr21_-_44898015 | 0.11 |
ENST00000332440.3 |
LINC00313 |
long intergenic non-protein coding RNA 313 |
| chr11_-_75062829 | 0.11 |
ENST00000393505.4 |
ARRB1 |
arrestin, beta 1 |
| chr12_+_7053172 | 0.11 |
ENST00000229281.5 |
C12orf57 |
chromosome 12 open reading frame 57 |
| chr2_-_191885686 | 0.11 |
ENST00000432058.1 |
STAT1 |
signal transducer and activator of transcription 1, 91kDa |
| chr1_+_24646002 | 0.11 |
ENST00000356046.2 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr1_+_24645807 | 0.11 |
ENST00000361548.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr6_+_80341000 | 0.11 |
ENST00000369838.4 |
SH3BGRL2 |
SH3 domain binding glutamic acid-rich protein like 2 |
| chr18_-_53068911 | 0.11 |
ENST00000537856.3 |
TCF4 |
transcription factor 4 |
| chr15_-_40600026 | 0.11 |
ENST00000456256.2 ENST00000557821.1 |
PLCB2 |
phospholipase C, beta 2 |
| chr8_+_356942 | 0.10 |
ENST00000276326.5 |
FBXO25 |
F-box protein 25 |
| chr11_-_75062730 | 0.10 |
ENST00000420843.2 ENST00000360025.3 |
ARRB1 |
arrestin, beta 1 |
| chr10_+_13142225 | 0.10 |
ENST00000378747.3 |
OPTN |
optineurin |
| chr12_+_7053228 | 0.10 |
ENST00000540506.2 |
C12orf57 |
chromosome 12 open reading frame 57 |
| chr1_+_209602771 | 0.10 |
ENST00000440276.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr12_+_7052974 | 0.10 |
ENST00000544681.1 ENST00000537087.1 |
C12orf57 |
chromosome 12 open reading frame 57 |
| chr17_-_21156578 | 0.10 |
ENST00000399011.2 ENST00000468196.1 |
C17orf103 |
chromosome 17 open reading frame 103 |
| chr7_+_76054224 | 0.10 |
ENST00000394857.3 |
ZP3 |
zona pellucida glycoprotein 3 (sperm receptor) |
| chr21_+_34775698 | 0.10 |
ENST00000381995.1 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr1_+_24645865 | 0.10 |
ENST00000342072.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr21_+_33245548 | 0.10 |
ENST00000270112.2 |
HUNK |
hormonally up-regulated Neu-associated kinase |
| chr19_+_45504688 | 0.09 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr2_+_61108771 | 0.09 |
ENST00000394479.3 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr10_+_13142075 | 0.09 |
ENST00000378757.2 ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN |
optineurin |
| chr8_-_72274095 | 0.09 |
ENST00000303824.7 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr12_+_100867694 | 0.09 |
ENST00000392986.3 ENST00000549996.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr9_+_139877445 | 0.09 |
ENST00000408973.2 |
LCNL1 |
lipocalin-like 1 |
| chr21_+_34775772 | 0.09 |
ENST00000405436.1 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr16_+_3096638 | 0.09 |
ENST00000336577.4 |
MMP25 |
matrix metallopeptidase 25 |
| chr8_-_72274467 | 0.09 |
ENST00000340726.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr5_+_175298573 | 0.09 |
ENST00000512824.1 |
CPLX2 |
complexin 2 |
| chr12_-_8815404 | 0.09 |
ENST00000359478.2 ENST00000396549.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr7_+_86273218 | 0.09 |
ENST00000361669.2 |
GRM3 |
glutamate receptor, metabotropic 3 |
| chr11_-_64052111 | 0.09 |
ENST00000394532.3 ENST00000394531.3 ENST00000309032.3 |
BAD |
BCL2-associated agonist of cell death |
| chr14_-_89883412 | 0.08 |
ENST00000557258.1 |
FOXN3 |
forkhead box N3 |
| chr14_+_91580708 | 0.08 |
ENST00000518868.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chrX_+_107683096 | 0.08 |
ENST00000328300.6 ENST00000361603.2 |
COL4A5 |
collagen, type IV, alpha 5 |
| chr14_-_101034407 | 0.08 |
ENST00000443071.2 ENST00000557378.1 |
BEGAIN |
brain-enriched guanylate kinase-associated |
| chr4_-_185395672 | 0.08 |
ENST00000393593.3 |
IRF2 |
interferon regulatory factor 2 |
| chr17_-_7590745 | 0.08 |
ENST00000514944.1 ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53 |
tumor protein p53 |
| chr1_-_146644036 | 0.08 |
ENST00000425272.2 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr13_-_78492927 | 0.08 |
ENST00000334286.5 |
EDNRB |
endothelin receptor type B |
| chr5_+_140579162 | 0.08 |
ENST00000536699.1 ENST00000354757.3 |
PCDHB11 |
protocadherin beta 11 |
| chr1_-_173886491 | 0.08 |
ENST00000367698.3 |
SERPINC1 |
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr16_+_67282853 | 0.08 |
ENST00000299798.11 |
SLC9A5 |
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr16_+_29973351 | 0.08 |
ENST00000602948.1 ENST00000279396.6 ENST00000575829.2 ENST00000561899.2 |
TMEM219 |
transmembrane protein 219 |
| chr13_+_32605437 | 0.08 |
ENST00000380250.3 |
FRY |
furry homolog (Drosophila) |
| chr20_+_9494987 | 0.08 |
ENST00000427562.2 ENST00000246070.2 |
LAMP5 |
lysosomal-associated membrane protein family, member 5 |
| chr6_+_292051 | 0.08 |
ENST00000344450.5 |
DUSP22 |
dual specificity phosphatase 22 |
| chr14_+_24583836 | 0.08 |
ENST00000559115.1 ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chr9_-_130639997 | 0.08 |
ENST00000373176.1 |
AK1 |
adenylate kinase 1 |
| chr12_+_7055631 | 0.08 |
ENST00000543115.1 ENST00000399448.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chrX_+_117861535 | 0.07 |
ENST00000371666.3 ENST00000371642.1 |
IL13RA1 |
interleukin 13 receptor, alpha 1 |
| chr18_+_21529811 | 0.07 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr11_+_63997750 | 0.07 |
ENST00000321685.3 |
DNAJC4 |
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr12_+_70760056 | 0.07 |
ENST00000258111.4 |
KCNMB4 |
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr1_-_153113927 | 0.07 |
ENST00000368752.4 |
SPRR2B |
small proline-rich protein 2B |
| chr8_+_21911054 | 0.07 |
ENST00000519850.1 ENST00000381470.3 |
DMTN |
dematin actin binding protein |
| chr2_+_64681103 | 0.07 |
ENST00000464281.1 |
LGALSL |
lectin, galactoside-binding-like |
| chr2_+_181845843 | 0.07 |
ENST00000602710.1 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
| chrX_-_24665208 | 0.07 |
ENST00000356768.4 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr19_-_50143452 | 0.07 |
ENST00000246792.3 |
RRAS |
related RAS viral (r-ras) oncogene homolog |
| chr10_-_105238997 | 0.07 |
ENST00000369783.4 |
CALHM3 |
calcium homeostasis modulator 3 |
| chr1_-_186649543 | 0.07 |
ENST00000367468.5 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr5_-_150460539 | 0.06 |
ENST00000520931.1 ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr15_+_26360970 | 0.06 |
ENST00000556159.1 ENST00000557523.1 |
LINC00929 |
long intergenic non-protein coding RNA 929 |
| chr12_-_86230315 | 0.06 |
ENST00000361228.3 |
RASSF9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr15_-_73661605 | 0.06 |
ENST00000261917.3 |
HCN4 |
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr10_+_18948311 | 0.06 |
ENST00000377275.3 |
ARL5B |
ADP-ribosylation factor-like 5B |
| chr9_+_4490394 | 0.06 |
ENST00000262352.3 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr18_+_44526786 | 0.06 |
ENST00000245121.5 ENST00000356157.7 |
KATNAL2 |
katanin p60 subunit A-like 2 |
| chr18_-_71959159 | 0.06 |
ENST00000494131.2 ENST00000397914.4 ENST00000340533.4 |
CYB5A |
cytochrome b5 type A (microsomal) |
| chr17_+_40440481 | 0.06 |
ENST00000590726.2 ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A |
signal transducer and activator of transcription 5A |
| chr17_-_45908875 | 0.06 |
ENST00000351111.2 ENST00000414011.1 |
MRPL10 |
mitochondrial ribosomal protein L10 |
| chr14_+_91580777 | 0.06 |
ENST00000525393.2 ENST00000428926.2 ENST00000517362.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr20_+_17207636 | 0.06 |
ENST00000262545.2 |
PCSK2 |
proprotein convertase subtilisin/kexin type 2 |
| chr10_+_73975742 | 0.06 |
ENST00000299381.4 |
ANAPC16 |
anaphase promoting complex subunit 16 |
| chr7_+_22766766 | 0.06 |
ENST00000426291.1 ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6 |
interleukin 6 (interferon, beta 2) |
| chr1_-_146644122 | 0.06 |
ENST00000254101.3 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr1_-_205744574 | 0.06 |
ENST00000367139.3 ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr11_-_124806297 | 0.06 |
ENST00000298251.4 |
HEPACAM |
hepatic and glial cell adhesion molecule |
| chr20_+_17207665 | 0.06 |
ENST00000536609.1 |
PCSK2 |
proprotein convertase subtilisin/kexin type 2 |
| chr21_+_34775181 | 0.06 |
ENST00000290219.6 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr5_-_150460914 | 0.06 |
ENST00000389378.2 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr17_-_61920280 | 0.06 |
ENST00000448276.2 ENST00000577990.1 |
SMARCD2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr6_-_11779840 | 0.06 |
ENST00000506810.1 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr2_-_204400013 | 0.06 |
ENST00000374489.2 ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1 |
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr12_+_50366620 | 0.06 |
ENST00000315520.5 |
AQP6 |
aquaporin 6, kidney specific |
| chr12_-_92539614 | 0.06 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr11_+_118478313 | 0.06 |
ENST00000356063.5 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
| chr9_+_75263565 | 0.06 |
ENST00000396237.3 |
TMC1 |
transmembrane channel-like 1 |
| chr14_+_91581011 | 0.06 |
ENST00000523894.1 ENST00000522322.1 ENST00000523771.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr19_-_33360647 | 0.05 |
ENST00000590341.1 ENST00000587772.1 ENST00000023064.4 |
SLC7A9 |
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr20_+_57875758 | 0.05 |
ENST00000395654.3 |
EDN3 |
endothelin 3 |
| chr8_+_22022800 | 0.05 |
ENST00000397814.3 |
BMP1 |
bone morphogenetic protein 1 |
| chr1_-_32229523 | 0.05 |
ENST00000398547.1 ENST00000373655.2 ENST00000373658.3 ENST00000257070.4 |
BAI2 |
brain-specific angiogenesis inhibitor 2 |
| chr1_+_43291220 | 0.05 |
ENST00000372514.3 |
ERMAP |
erythroblast membrane-associated protein (Scianna blood group) |
| chr11_+_101785727 | 0.05 |
ENST00000263468.8 |
KIAA1377 |
KIAA1377 |
| chr16_+_50730910 | 0.05 |
ENST00000300589.2 |
NOD2 |
nucleotide-binding oligomerization domain containing 2 |
| chr14_+_23299088 | 0.05 |
ENST00000355151.5 ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52 |
mitochondrial ribosomal protein L52 |
| chr16_-_67978016 | 0.05 |
ENST00000264005.5 |
LCAT |
lecithin-cholesterol acyltransferase |
| chr22_+_23134974 | 0.05 |
ENST00000390314.2 |
IGLV2-11 |
immunoglobulin lambda variable 2-11 |
| chr3_-_113775328 | 0.05 |
ENST00000483766.1 ENST00000545063.1 ENST00000491000.1 ENST00000295878.3 |
KIAA1407 |
KIAA1407 |
| chr9_-_125667618 | 0.05 |
ENST00000423239.2 |
RC3H2 |
ring finger and CCCH-type domains 2 |
| chr10_+_13141585 | 0.05 |
ENST00000378764.2 |
OPTN |
optineurin |
| chr17_-_40540377 | 0.05 |
ENST00000404395.3 ENST00000389272.3 ENST00000585517.1 ENST00000588065.1 |
STAT3 |
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr1_-_12677714 | 0.05 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chr1_+_54359854 | 0.05 |
ENST00000361921.3 ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1 |
deiodinase, iodothyronine, type I |
| chr16_+_2059872 | 0.05 |
ENST00000567649.1 |
NPW |
neuropeptide W |
| chr17_-_40540484 | 0.05 |
ENST00000588969.1 |
STAT3 |
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr11_+_57508825 | 0.05 |
ENST00000534355.1 |
C11orf31 |
chromosome 11 open reading frame 31 |
| chr14_-_30396948 | 0.05 |
ENST00000331968.5 |
PRKD1 |
protein kinase D1 |
| chr1_+_24117627 | 0.05 |
ENST00000400061.1 |
LYPLA2 |
lysophospholipase II |
| chr1_+_110453203 | 0.05 |
ENST00000357302.4 ENST00000344188.5 ENST00000329608.6 |
CSF1 |
colony stimulating factor 1 (macrophage) |
| chr11_+_64052266 | 0.05 |
ENST00000539851.1 |
GPR137 |
G protein-coupled receptor 137 |
| chr6_+_36165133 | 0.05 |
ENST00000446974.1 ENST00000454960.1 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
| chr14_+_88852059 | 0.05 |
ENST00000045347.7 |
SPATA7 |
spermatogenesis associated 7 |
| chr4_-_74864386 | 0.05 |
ENST00000296027.4 |
CXCL5 |
chemokine (C-X-C motif) ligand 5 |
| chrX_-_134186144 | 0.05 |
ENST00000370775.2 |
FAM127B |
family with sequence similarity 127, member B |
| chr1_+_8021954 | 0.05 |
ENST00000377491.1 ENST00000377488.1 |
PARK7 |
parkinson protein 7 |
| chr10_+_22605304 | 0.05 |
ENST00000475460.2 ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1 COMMD3 |
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr3_-_107941230 | 0.05 |
ENST00000264538.3 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr12_-_7818474 | 0.05 |
ENST00000229304.4 |
APOBEC1 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr10_+_49514698 | 0.05 |
ENST00000432379.1 ENST00000429041.1 ENST00000374189.1 |
MAPK8 |
mitogen-activated protein kinase 8 |
| chr1_+_151020175 | 0.05 |
ENST00000368926.5 |
C1orf56 |
chromosome 1 open reading frame 56 |
| chr5_+_140588269 | 0.05 |
ENST00000541609.1 ENST00000239450.2 |
PCDHB12 |
protocadherin beta 12 |
| chr1_+_37940153 | 0.04 |
ENST00000373087.6 |
ZC3H12A |
zinc finger CCCH-type containing 12A |
| chr10_-_102090243 | 0.04 |
ENST00000338519.3 ENST00000353274.3 ENST00000318222.3 |
PKD2L1 |
polycystic kidney disease 2-like 1 |
| chr1_+_156611704 | 0.04 |
ENST00000329117.5 |
BCAN |
brevican |
| chr20_-_32891151 | 0.04 |
ENST00000217426.2 |
AHCY |
adenosylhomocysteinase |
| chr9_-_32526184 | 0.04 |
ENST00000545044.1 ENST00000379868.1 |
DDX58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr22_-_38380543 | 0.04 |
ENST00000396884.2 |
SOX10 |
SRY (sex determining region Y)-box 10 |
| chr11_+_64052454 | 0.04 |
ENST00000539833.1 |
GPR137 |
G protein-coupled receptor 137 |
| chr2_+_113735575 | 0.04 |
ENST00000376489.2 ENST00000259205.4 |
IL36G |
interleukin 36, gamma |
| chr19_+_14142535 | 0.04 |
ENST00000263379.2 |
IL27RA |
interleukin 27 receptor, alpha |
| chr10_+_22605374 | 0.04 |
ENST00000448361.1 |
COMMD3 |
COMM domain containing 3 |
| chr2_+_166428839 | 0.04 |
ENST00000342316.4 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
| chr8_-_16859690 | 0.04 |
ENST00000180166.5 |
FGF20 |
fibroblast growth factor 20 |
| chr14_+_24701628 | 0.04 |
ENST00000355299.4 ENST00000559836.1 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr9_-_32526299 | 0.04 |
ENST00000379882.1 ENST00000379883.2 |
DDX58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chrX_+_108780347 | 0.04 |
ENST00000372103.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr12_-_15114191 | 0.04 |
ENST00000541380.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr9_-_123476719 | 0.04 |
ENST00000373930.3 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr2_+_208394455 | 0.04 |
ENST00000430624.1 |
CREB1 |
cAMP responsive element binding protein 1 |
| chr11_-_61560053 | 0.04 |
ENST00000537328.1 |
TMEM258 |
transmembrane protein 258 |
| chr7_-_111202511 | 0.04 |
ENST00000452895.1 ENST00000452753.1 ENST00000331762.3 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr2_+_74154032 | 0.04 |
ENST00000356837.6 |
DGUOK |
deoxyguanosine kinase |
| chr11_+_64052692 | 0.04 |
ENST00000377702.4 |
GPR137 |
G protein-coupled receptor 137 |
| chrX_+_108779870 | 0.04 |
ENST00000372107.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chrX_+_153059608 | 0.04 |
ENST00000370087.1 |
SSR4 |
signal sequence receptor, delta |
| chr19_-_39390440 | 0.04 |
ENST00000249396.7 ENST00000414941.1 ENST00000392081.2 |
SIRT2 |
sirtuin 2 |
| chr11_+_74951948 | 0.04 |
ENST00000562197.2 |
TPBGL |
trophoblast glycoprotein-like |
| chrX_+_108780062 | 0.04 |
ENST00000372106.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr7_+_95401877 | 0.04 |
ENST00000524053.1 ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
| chr10_+_70661014 | 0.04 |
ENST00000373585.3 |
DDX50 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr9_-_123476612 | 0.04 |
ENST00000426959.1 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr11_+_64052294 | 0.04 |
ENST00000536667.1 |
GPR137 |
G protein-coupled receptor 137 |
| chr19_-_38747172 | 0.04 |
ENST00000347262.4 ENST00000591585.1 ENST00000301242.4 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr19_-_39390350 | 0.04 |
ENST00000447739.1 ENST00000358931.5 ENST00000407552.1 |
SIRT2 |
sirtuin 2 |
| chr2_-_89619904 | 0.04 |
ENST00000498574.1 |
IGKV1-39 |
immunoglobulin kappa variable 1-39 (gene/pseudogene) |
| chrX_+_154610428 | 0.04 |
ENST00000354514.4 |
H2AFB2 |
H2A histone family, member B2 |
| chr15_-_82824843 | 0.04 |
ENST00000560826.1 ENST00000559187.1 ENST00000330339.7 |
RPS17 |
ribosomal protein S17 |
| chr2_+_74153953 | 0.04 |
ENST00000264093.4 ENST00000348222.1 |
DGUOK |
deoxyguanosine kinase |
| chr6_-_32636145 | 0.04 |
ENST00000399084.1 |
HLA-DQB1 |
major histocompatibility complex, class II, DQ beta 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.3 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0001079 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.3 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:1903094 | enzyme active site formation(GO:0018307) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) detoxification of mercury ion(GO:0050787) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.3 | GO:1904417 | regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.0 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.3 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.0 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.1 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0090107 | regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.0 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.1 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.0 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.3 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.2 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.0 | 0.2 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |