Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
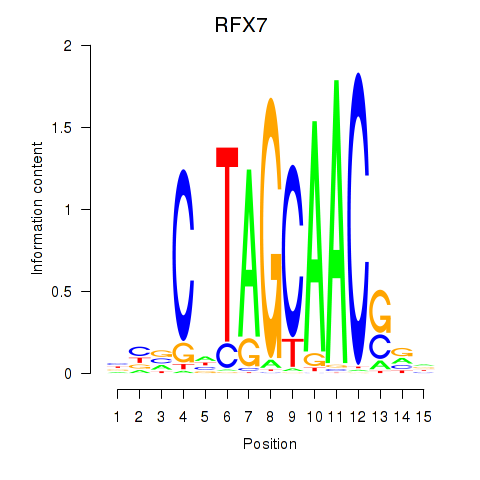
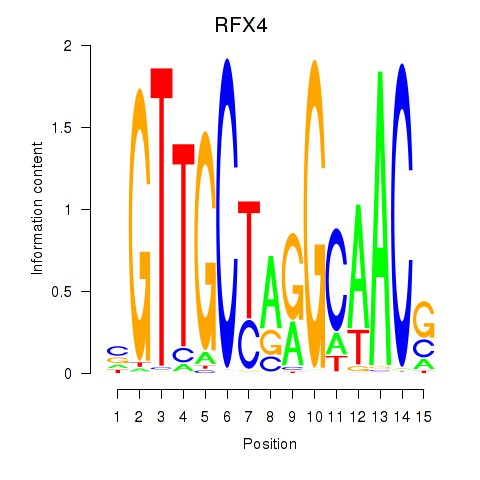
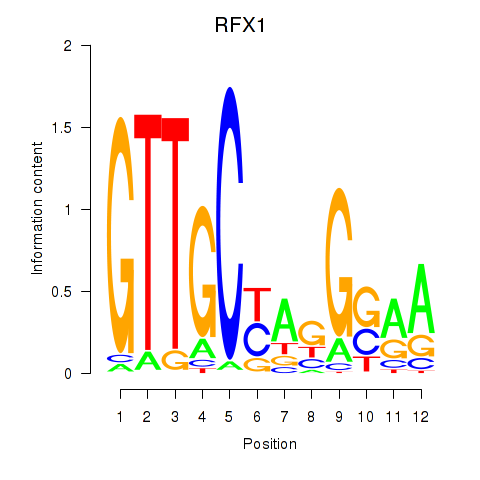
Results for RFX7_RFX4_RFX1
Z-value: 0.85
Transcription factors associated with RFX7_RFX4_RFX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX7
|
ENSG00000181827.10 | RFX7 |
|
RFX4
|
ENSG00000111783.8 | RFX4 |
|
RFX1
|
ENSG00000132005.4 | RFX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX7 | hg19_v2_chr15_-_56535464_56535521 | -0.61 | 1.1e-01 | Click! |
| RFX1 | hg19_v2_chr19_-_14117074_14117141 | -0.39 | 3.5e-01 | Click! |
| RFX4 | hg19_v2_chr12_+_106976678_106976708, hg19_v2_chr12_+_107078474_107078533 | -0.01 | 9.8e-01 | Click! |
Activity profile of RFX7_RFX4_RFX1 motif
Sorted Z-values of RFX7_RFX4_RFX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX7_RFX4_RFX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_50316517 | 0.66 |
ENST00000313777.4 ENST00000445575.2 |
FUZ |
fuzzy planar cell polarity protein |
| chr19_-_50316423 | 0.62 |
ENST00000528094.1 ENST00000526575.1 |
FUZ |
fuzzy planar cell polarity protein |
| chr19_-_50316489 | 0.60 |
ENST00000533418.1 |
FUZ |
fuzzy planar cell polarity protein |
| chr1_-_72748417 | 0.53 |
ENST00000357731.5 |
NEGR1 |
neuronal growth regulator 1 |
| chr2_-_190044480 | 0.46 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr1_-_109655355 | 0.41 |
ENST00000369945.3 |
C1orf194 |
chromosome 1 open reading frame 194 |
| chr14_-_61116168 | 0.41 |
ENST00000247182.6 |
SIX1 |
SIX homeobox 1 |
| chr20_-_30310797 | 0.40 |
ENST00000422920.1 |
BCL2L1 |
BCL2-like 1 |
| chr11_-_6426635 | 0.40 |
ENST00000608645.1 ENST00000608394.1 ENST00000529519.1 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr20_-_30311703 | 0.38 |
ENST00000450273.1 ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1 |
BCL2-like 1 |
| chr19_+_48972459 | 0.34 |
ENST00000427476.1 |
CYTH2 |
cytohesin 2 |
| chr11_-_70507901 | 0.33 |
ENST00000449833.2 ENST00000357171.3 ENST00000449116.2 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
| chr19_-_45953983 | 0.32 |
ENST00000592083.1 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr17_-_15244894 | 0.30 |
ENST00000338696.2 ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3 |
tektin 3 |
| chr2_+_95537170 | 0.28 |
ENST00000295201.4 |
TEKT4 |
tektin 4 |
| chr2_-_62081148 | 0.25 |
ENST00000404929.1 |
FAM161A |
family with sequence similarity 161, member A |
| chr2_-_62081254 | 0.25 |
ENST00000405894.3 |
FAM161A |
family with sequence similarity 161, member A |
| chr2_-_63815628 | 0.24 |
ENST00000409562.3 |
WDPCP |
WD repeat containing planar cell polarity effector |
| chr20_-_44718538 | 0.24 |
ENST00000290231.6 ENST00000372291.3 |
NCOA5 |
nuclear receptor coactivator 5 |
| chr22_-_43485381 | 0.23 |
ENST00000331018.7 ENST00000266254.7 ENST00000445824.1 |
TTLL1 |
tubulin tyrosine ligase-like family, member 1 |
| chr3_-_128712906 | 0.23 |
ENST00000511438.1 |
KIAA1257 |
KIAA1257 |
| chr19_+_55996565 | 0.23 |
ENST00000587400.1 |
NAT14 |
N-acetyltransferase 14 (GCN5-related, putative) |
| chr14_-_88459182 | 0.22 |
ENST00000544807.2 |
GALC |
galactosylceramidase |
| chr4_+_26585686 | 0.22 |
ENST00000505206.1 ENST00000511789.1 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr4_-_2420335 | 0.22 |
ENST00000503000.1 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
| chr9_-_138391692 | 0.22 |
ENST00000429260.2 |
C9orf116 |
chromosome 9 open reading frame 116 |
| chr10_+_25305524 | 0.22 |
ENST00000524413.1 ENST00000376356.4 |
THNSL1 |
threonine synthase-like 1 (S. cerevisiae) |
| chr1_+_183605200 | 0.22 |
ENST00000304685.4 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr11_-_70507867 | 0.21 |
ENST00000412252.1 ENST00000409161.1 ENST00000409530.1 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
| chr19_-_40324255 | 0.21 |
ENST00000593685.1 ENST00000600611.1 |
DYRK1B |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr19_+_16308659 | 0.21 |
ENST00000590263.1 ENST00000590756.1 ENST00000541844.1 |
AP1M1 |
adaptor-related protein complex 1, mu 1 subunit |
| chr4_+_26585538 | 0.21 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr16_-_72127456 | 0.21 |
ENST00000562153.1 |
TXNL4B |
thioredoxin-like 4B |
| chr14_-_88459503 | 0.20 |
ENST00000393568.4 ENST00000261304.2 |
GALC |
galactosylceramidase |
| chr1_+_155829286 | 0.20 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr19_-_51014460 | 0.20 |
ENST00000595669.1 |
JOSD2 |
Josephin domain containing 2 |
| chr3_+_99357319 | 0.20 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr12_-_76478686 | 0.20 |
ENST00000261182.8 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr1_-_109655377 | 0.19 |
ENST00000369948.3 |
C1orf194 |
chromosome 1 open reading frame 194 |
| chr15_+_55700741 | 0.18 |
ENST00000569691.1 |
C15orf65 |
chromosome 15 open reading frame 65 |
| chr22_+_35776828 | 0.18 |
ENST00000216117.8 |
HMOX1 |
heme oxygenase (decycling) 1 |
| chr8_+_13424352 | 0.18 |
ENST00000297324.4 |
C8orf48 |
chromosome 8 open reading frame 48 |
| chr22_-_32555275 | 0.18 |
ENST00000382097.3 |
C22orf42 |
chromosome 22 open reading frame 42 |
| chr19_+_55996316 | 0.18 |
ENST00000205194.4 |
NAT14 |
N-acetyltransferase 14 (GCN5-related, putative) |
| chr20_+_12989596 | 0.18 |
ENST00000434210.1 ENST00000399002.2 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
| chr17_-_64187973 | 0.17 |
ENST00000583358.1 ENST00000392769.2 |
CEP112 |
centrosomal protein 112kDa |
| chr17_-_64188177 | 0.17 |
ENST00000535342.2 |
CEP112 |
centrosomal protein 112kDa |
| chr12_-_58329819 | 0.17 |
ENST00000551421.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr11_+_71791693 | 0.17 |
ENST00000289488.2 ENST00000447974.1 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
| chr6_+_89790459 | 0.17 |
ENST00000369472.1 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr2_-_63815860 | 0.16 |
ENST00000272321.7 ENST00000431065.1 |
WDPCP |
WD repeat containing planar cell polarity effector |
| chr8_+_10530155 | 0.16 |
ENST00000521818.1 |
C8orf74 |
chromosome 8 open reading frame 74 |
| chr2_+_170550944 | 0.16 |
ENST00000359744.3 ENST00000438838.1 ENST00000438710.1 ENST00000449906.1 ENST00000498202.2 ENST00000272797.4 |
PHOSPHO2 KLHL23 |
phosphatase, orphan 2 kelch-like family member 23 |
| chr6_-_167275991 | 0.16 |
ENST00000510118.1 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr20_+_17550489 | 0.16 |
ENST00000246069.7 |
DSTN |
destrin (actin depolymerizing factor) |
| chr5_+_75699149 | 0.16 |
ENST00000379730.3 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr19_+_507299 | 0.16 |
ENST00000359315.5 |
TPGS1 |
tubulin polyglutamylase complex subunit 1 |
| chr4_-_156787425 | 0.15 |
ENST00000537611.2 |
ASIC5 |
acid-sensing (proton-gated) ion channel family member 5 |
| chr14_-_22005018 | 0.15 |
ENST00000546363.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr19_-_46974664 | 0.15 |
ENST00000438932.2 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr1_-_85155939 | 0.15 |
ENST00000603677.1 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chrX_-_64254587 | 0.15 |
ENST00000337990.2 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr19_-_46974741 | 0.15 |
ENST00000313683.10 ENST00000602246.1 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr6_-_167276033 | 0.15 |
ENST00000503859.1 ENST00000506565.1 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr16_+_67700673 | 0.15 |
ENST00000403458.4 ENST00000602365.1 |
C16orf86 |
chromosome 16 open reading frame 86 |
| chr16_+_20912382 | 0.15 |
ENST00000396052.2 |
LYRM1 |
LYR motif containing 1 |
| chr17_-_56296580 | 0.15 |
ENST00000313863.6 ENST00000546108.1 ENST00000337050.7 ENST00000393119.2 |
MKS1 |
Meckel syndrome, type 1 |
| chr1_-_85156216 | 0.15 |
ENST00000342203.3 ENST00000370612.4 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr1_-_85156090 | 0.15 |
ENST00000605755.1 ENST00000437941.2 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr15_-_55790515 | 0.15 |
ENST00000448430.2 ENST00000457155.2 |
DYX1C1 |
dyslexia susceptibility 1 candidate 1 |
| chr3_-_105588231 | 0.15 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr19_-_40324767 | 0.14 |
ENST00000601972.1 ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr10_+_15001430 | 0.14 |
ENST00000407572.1 |
MEIG1 |
meiosis/spermiogenesis associated 1 |
| chr16_+_20912075 | 0.14 |
ENST00000219168.4 |
LYRM1 |
LYR motif containing 1 |
| chr17_+_48585794 | 0.14 |
ENST00000576179.1 ENST00000419930.1 |
MYCBPAP |
MYCBP associated protein |
| chr17_+_42977122 | 0.14 |
ENST00000412523.2 ENST00000331733.4 ENST00000417826.2 |
FAM187A CCDC103 |
family with sequence similarity 187, member A coiled-coil domain containing 103 |
| chr12_+_107168418 | 0.14 |
ENST00000392839.2 ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
| chr1_-_36915880 | 0.14 |
ENST00000445843.3 |
OSCP1 |
organic solute carrier partner 1 |
| chr14_+_61447927 | 0.14 |
ENST00000451406.1 |
SLC38A6 |
solute carrier family 38, member 6 |
| chr11_+_133938820 | 0.14 |
ENST00000299106.4 ENST00000529443.2 |
JAM3 |
junctional adhesion molecule 3 |
| chr16_+_3550924 | 0.13 |
ENST00000576634.1 ENST00000574369.1 ENST00000341633.5 ENST00000417763.2 ENST00000571025.1 |
CLUAP1 |
clusterin associated protein 1 |
| chr1_-_36916066 | 0.13 |
ENST00000315643.9 |
OSCP1 |
organic solute carrier partner 1 |
| chr11_+_71791359 | 0.13 |
ENST00000419228.1 ENST00000435085.1 ENST00000307198.7 ENST00000538413.1 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
| chr14_+_61447832 | 0.13 |
ENST00000354886.2 ENST00000267488.4 |
SLC38A6 |
solute carrier family 38, member 6 |
| chrX_+_102883620 | 0.13 |
ENST00000372626.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr11_-_62521614 | 0.13 |
ENST00000527994.1 ENST00000394807.3 |
ZBTB3 |
zinc finger and BTB domain containing 3 |
| chr13_+_24153488 | 0.13 |
ENST00000382258.4 ENST00000382263.3 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr2_+_242089046 | 0.13 |
ENST00000407025.1 ENST00000272983.8 |
PPP1R7 |
protein phosphatase 1, regulatory subunit 7 |
| chr2_+_242088963 | 0.13 |
ENST00000438799.1 ENST00000402734.1 ENST00000423280.1 |
PPP1R7 |
protein phosphatase 1, regulatory subunit 7 |
| chr4_+_6784401 | 0.13 |
ENST00000425103.1 ENST00000307659.5 |
KIAA0232 |
KIAA0232 |
| chr4_+_153457404 | 0.13 |
ENST00000604157.1 ENST00000594836.1 |
MIR4453 |
microRNA 4453 |
| chr1_-_109968973 | 0.13 |
ENST00000271308.4 ENST00000538610.1 |
PSMA5 |
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr11_-_66104237 | 0.12 |
ENST00000530056.1 |
RIN1 |
Ras and Rab interactor 1 |
| chr4_+_15004165 | 0.12 |
ENST00000538197.1 ENST00000541112.1 ENST00000442003.2 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
| chr1_+_161068179 | 0.12 |
ENST00000368011.4 ENST00000392192.2 |
KLHDC9 |
kelch domain containing 9 |
| chr1_+_39456895 | 0.12 |
ENST00000432648.3 ENST00000446189.2 ENST00000372984.4 |
AKIRIN1 |
akirin 1 |
| chr11_+_133938955 | 0.12 |
ENST00000534549.1 ENST00000441717.3 |
JAM3 |
junctional adhesion molecule 3 |
| chr4_-_89619386 | 0.12 |
ENST00000323061.5 |
NAP1L5 |
nucleosome assembly protein 1-like 5 |
| chr1_+_11866207 | 0.12 |
ENST00000312413.6 ENST00000346436.6 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr11_+_71791803 | 0.12 |
ENST00000539271.1 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
| chr6_+_112408768 | 0.12 |
ENST00000368656.2 ENST00000604268.1 |
FAM229B |
family with sequence similarity 229, member B |
| chr2_-_175869936 | 0.12 |
ENST00000409900.3 |
CHN1 |
chimerin 1 |
| chr4_-_2420357 | 0.12 |
ENST00000511071.1 ENST00000509171.1 ENST00000290974.2 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
| chr1_-_36916011 | 0.12 |
ENST00000356637.5 ENST00000354267.3 ENST00000235532.5 |
OSCP1 |
organic solute carrier partner 1 |
| chr20_-_45981138 | 0.12 |
ENST00000446994.2 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr2_+_61293021 | 0.12 |
ENST00000402291.1 |
KIAA1841 |
KIAA1841 |
| chr6_-_44265446 | 0.11 |
ENST00000371503.3 |
TCTE1 |
t-complex-associated-testis-expressed 1 |
| chr7_+_158649242 | 0.11 |
ENST00000407559.3 |
WDR60 |
WD repeat domain 60 |
| chr15_-_55700216 | 0.11 |
ENST00000569205.1 |
CCPG1 |
cell cycle progression 1 |
| chr5_+_178986693 | 0.11 |
ENST00000437570.2 ENST00000393438.2 |
RUFY1 |
RUN and FYVE domain containing 1 |
| chr15_-_55700457 | 0.11 |
ENST00000442196.3 ENST00000563171.1 ENST00000425574.3 |
CCPG1 |
cell cycle progression 1 |
| chr3_+_191046810 | 0.11 |
ENST00000392455.3 ENST00000392456.3 |
CCDC50 |
coiled-coil domain containing 50 |
| chr2_-_230579185 | 0.11 |
ENST00000341772.4 |
DNER |
delta/notch-like EGF repeat containing |
| chr6_+_52285131 | 0.11 |
ENST00000433625.2 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
| chr6_-_38607673 | 0.11 |
ENST00000481247.1 |
BTBD9 |
BTB (POZ) domain containing 9 |
| chrX_+_135251835 | 0.11 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr2_-_175870085 | 0.10 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr13_-_44361025 | 0.10 |
ENST00000261488.6 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
| chr12_-_54867352 | 0.10 |
ENST00000305879.5 |
GTSF1 |
gametocyte specific factor 1 |
| chr20_+_42219559 | 0.10 |
ENST00000373030.3 ENST00000373039.4 |
IFT52 |
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr12_-_49582978 | 0.10 |
ENST00000301071.7 |
TUBA1A |
tubulin, alpha 1a |
| chr12_-_76478446 | 0.10 |
ENST00000393263.3 ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr12_-_76478417 | 0.10 |
ENST00000552342.1 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr9_-_130635741 | 0.10 |
ENST00000223836.10 |
AK1 |
adenylate kinase 1 |
| chr15_+_93447675 | 0.10 |
ENST00000536619.1 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr19_+_46850251 | 0.10 |
ENST00000012443.4 |
PPP5C |
protein phosphatase 5, catalytic subunit |
| chr19_+_16308711 | 0.10 |
ENST00000429941.2 ENST00000444449.2 ENST00000589822.1 |
AP1M1 |
adaptor-related protein complex 1, mu 1 subunit |
| chr4_-_170533723 | 0.10 |
ENST00000510533.1 ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1 |
NIMA-related kinase 1 |
| chr20_+_17550691 | 0.10 |
ENST00000474024.1 |
DSTN |
destrin (actin depolymerizing factor) |
| chr1_-_159893507 | 0.10 |
ENST00000368096.1 |
TAGLN2 |
transgelin 2 |
| chr1_+_144220127 | 0.10 |
ENST00000369373.5 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr5_-_133702761 | 0.10 |
ENST00000521118.1 ENST00000265334.4 ENST00000435211.1 |
CDKL3 |
cyclin-dependent kinase-like 3 |
| chr11_+_69455855 | 0.10 |
ENST00000227507.2 ENST00000536559.1 |
CCND1 |
cyclin D1 |
| chr14_+_88852059 | 0.09 |
ENST00000045347.7 |
SPATA7 |
spermatogenesis associated 7 |
| chr20_+_306177 | 0.09 |
ENST00000544632.1 |
SOX12 |
SRY (sex determining region Y)-box 12 |
| chr12_+_107168342 | 0.09 |
ENST00000392837.4 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
| chr7_+_12726474 | 0.09 |
ENST00000396662.1 ENST00000356797.3 ENST00000396664.2 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chrX_+_85969626 | 0.09 |
ENST00000484479.1 |
DACH2 |
dachshund homolog 2 (Drosophila) |
| chr11_+_6411670 | 0.09 |
ENST00000530395.1 ENST00000527275.1 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr10_-_28623368 | 0.09 |
ENST00000441595.2 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr12_-_76478386 | 0.09 |
ENST00000535020.2 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr16_+_284788 | 0.09 |
ENST00000301679.2 ENST00000419173.1 ENST00000426695.1 ENST00000438220.1 ENST00000447499.1 ENST00000453430.1 ENST00000442458.2 ENST00000449945.1 ENST00000420046.1 ENST00000301678.3 ENST00000450082.2 ENST00000420500.1 |
ITFG3 |
integrin alpha FG-GAP repeat containing 3 |
| chr15_-_55700522 | 0.09 |
ENST00000564092.1 ENST00000310958.6 |
CCPG1 |
cell cycle progression 1 |
| chr19_+_1249869 | 0.09 |
ENST00000591446.2 |
MIDN |
midnolin |
| chr22_+_45809560 | 0.09 |
ENST00000342894.3 ENST00000538017.1 |
RIBC2 |
RIB43A domain with coiled-coils 2 |
| chr6_+_52285046 | 0.09 |
ENST00000371068.5 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
| chr12_+_48577366 | 0.09 |
ENST00000316554.3 |
C12orf68 |
chromosome 12 open reading frame 68 |
| chr3_-_197676740 | 0.09 |
ENST00000452735.1 ENST00000453254.1 ENST00000455191.1 |
IQCG |
IQ motif containing G |
| chr19_+_46850320 | 0.09 |
ENST00000391919.1 |
PPP5C |
protein phosphatase 5, catalytic subunit |
| chr19_-_10679644 | 0.09 |
ENST00000393599.2 |
CDKN2D |
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr2_+_220110177 | 0.09 |
ENST00000409638.3 ENST00000396738.2 ENST00000409516.3 |
STK16 |
serine/threonine kinase 16 |
| chr3_+_9958758 | 0.09 |
ENST00000383812.4 ENST00000438091.1 ENST00000295981.3 ENST00000436503.1 ENST00000403601.3 ENST00000416074.2 ENST00000455057.1 |
IL17RC |
interleukin 17 receptor C |
| chr19_-_6393216 | 0.09 |
ENST00000595047.1 |
GTF2F1 |
general transcription factor IIF, polypeptide 1, 74kDa |
| chr17_-_55911970 | 0.09 |
ENST00000581805.1 ENST00000580960.1 |
RP11-60A24.3 |
RP11-60A24.3 |
| chr3_-_105587879 | 0.09 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr15_-_52944231 | 0.09 |
ENST00000546305.2 |
FAM214A |
family with sequence similarity 214, member A |
| chr6_-_130031358 | 0.09 |
ENST00000368149.2 |
ARHGAP18 |
Rho GTPase activating protein 18 |
| chr1_+_211432775 | 0.09 |
ENST00000419091.2 |
RCOR3 |
REST corepressor 3 |
| chr2_+_173940163 | 0.08 |
ENST00000539448.1 |
MLTK |
Mitogen-activated protein kinase kinase kinase MLT |
| chr17_-_16472483 | 0.08 |
ENST00000395824.1 ENST00000448349.2 ENST00000395825.3 |
ZNF287 |
zinc finger protein 287 |
| chrX_+_135252050 | 0.08 |
ENST00000449474.1 ENST00000345434.3 |
FHL1 |
four and a half LIM domains 1 |
| chrX_+_47444613 | 0.08 |
ENST00000445623.1 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
| chr2_-_153032484 | 0.08 |
ENST00000263904.4 |
STAM2 |
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr12_-_105630016 | 0.08 |
ENST00000258530.3 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr6_+_89790490 | 0.08 |
ENST00000336032.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr15_+_49447947 | 0.08 |
ENST00000327171.3 ENST00000560654.1 |
GALK2 |
galactokinase 2 |
| chr16_-_29757272 | 0.08 |
ENST00000329410.3 |
C16orf54 |
chromosome 16 open reading frame 54 |
| chr8_+_144816303 | 0.08 |
ENST00000533004.1 |
FAM83H-AS1 |
FAM83H antisense RNA 1 (head to head) |
| chr4_-_152149033 | 0.08 |
ENST00000514152.1 |
SH3D19 |
SH3 domain containing 19 |
| chr2_+_220144052 | 0.08 |
ENST00000425450.1 ENST00000392086.4 ENST00000421532.1 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr16_+_16429787 | 0.08 |
ENST00000331436.4 ENST00000541593.1 |
AC138969.4 |
Protein PKD1P1 |
| chrX_+_135251783 | 0.08 |
ENST00000394153.2 |
FHL1 |
four and a half LIM domains 1 |
| chr3_+_185304059 | 0.08 |
ENST00000427465.2 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chrX_-_131623874 | 0.08 |
ENST00000436215.1 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr6_-_166796461 | 0.08 |
ENST00000360961.6 ENST00000341756.6 |
MPC1 |
mitochondrial pyruvate carrier 1 |
| chr22_+_23487513 | 0.08 |
ENST00000263116.2 ENST00000341989.4 |
RAB36 |
RAB36, member RAS oncogene family |
| chr19_-_58951496 | 0.08 |
ENST00000254166.3 |
ZNF132 |
zinc finger protein 132 |
| chr11_+_6411636 | 0.08 |
ENST00000299397.3 ENST00000356761.2 ENST00000342245.4 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr12_+_4671352 | 0.08 |
ENST00000542744.1 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr11_-_66103932 | 0.08 |
ENST00000311320.4 |
RIN1 |
Ras and Rab interactor 1 |
| chr8_-_101118185 | 0.08 |
ENST00000523437.1 |
RGS22 |
regulator of G-protein signaling 22 |
| chr1_-_45140074 | 0.08 |
ENST00000420706.1 ENST00000372235.3 ENST00000372242.3 ENST00000372243.3 ENST00000372244.3 |
TMEM53 |
transmembrane protein 53 |
| chr3_-_10149907 | 0.08 |
ENST00000450660.2 ENST00000524279.1 |
FANCD2OS |
FANCD2 opposite strand |
| chr11_-_66103867 | 0.08 |
ENST00000424433.2 |
RIN1 |
Ras and Rab interactor 1 |
| chr2_+_48667983 | 0.08 |
ENST00000449090.2 |
PPP1R21 |
protein phosphatase 1, regulatory subunit 21 |
| chr7_+_156902674 | 0.07 |
ENST00000594086.1 |
AC006967.1 |
Protein LOC100996426 |
| chr8_+_22428457 | 0.07 |
ENST00000517962.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr15_+_85923856 | 0.07 |
ENST00000560302.1 ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr5_+_154237778 | 0.07 |
ENST00000523698.1 ENST00000517876.1 ENST00000520472.1 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr9_+_131218408 | 0.07 |
ENST00000351030.3 ENST00000604420.1 ENST00000535026.1 ENST00000448249.3 ENST00000393527.3 |
ODF2 |
outer dense fiber of sperm tails 2 |
| chr12_-_88535747 | 0.07 |
ENST00000309041.7 |
CEP290 |
centrosomal protein 290kDa |
| chr17_+_72270429 | 0.07 |
ENST00000311014.6 |
DNAI2 |
dynein, axonemal, intermediate chain 2 |
| chr20_+_30865429 | 0.07 |
ENST00000375712.3 |
KIF3B |
kinesin family member 3B |
| chr12_+_111051832 | 0.07 |
ENST00000550703.2 ENST00000551590.1 |
TCTN1 |
tectonic family member 1 |
| chr1_+_245133062 | 0.07 |
ENST00000366523.1 |
EFCAB2 |
EF-hand calcium binding domain 2 |
| chr10_+_35416090 | 0.07 |
ENST00000354759.3 |
CREM |
cAMP responsive element modulator |
| chr7_-_139477500 | 0.07 |
ENST00000406875.3 ENST00000428878.2 |
HIPK2 |
homeodomain interacting protein kinase 2 |
| chr2_+_26624775 | 0.07 |
ENST00000288710.2 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chr6_-_44923160 | 0.07 |
ENST00000371458.1 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
| chr12_+_53693466 | 0.07 |
ENST00000267103.5 ENST00000548632.1 |
C12orf10 |
chromosome 12 open reading frame 10 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.2 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.4 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.3 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.8 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.2 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 0.4 | GO:0090521 | regulation of embryonic cell shape(GO:0016476) septin cytoskeleton organization(GO:0032185) glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.2 | GO:1905154 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.1 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.1 | 0.5 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.4 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0006756 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.4 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.1 | GO:0071878 | negative regulation of adrenergic receptor signaling pathway(GO:0071878) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.0 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.1 | GO:2000370 | positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.3 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0018307 | tRNA wobble position uridine thiolation(GO:0002143) enzyme active site formation(GO:0018307) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0034125 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) |
| 0.0 | 0.0 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.0 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.2 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.0 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.0 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0016235 | inclusion body(GO:0016234) aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.4 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.0 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0044594 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |