Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
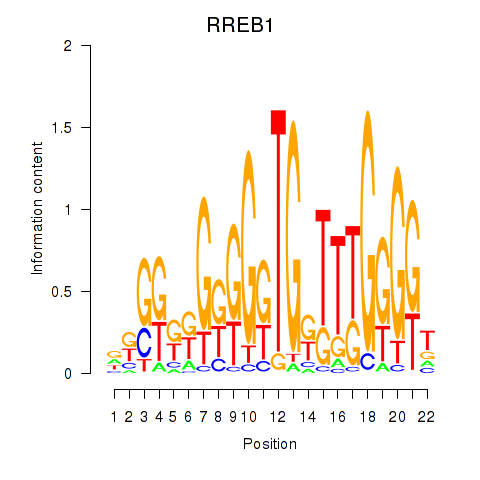
Results for RREB1
Z-value: 0.70
Transcription factors associated with RREB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RREB1
|
ENSG00000124782.15 | RREB1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RREB1 | hg19_v2_chr6_+_7107999_7108054 | 0.71 | 4.8e-02 | Click! |
Activity profile of RREB1 motif
Sorted Z-values of RREB1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RREB1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_35609380 | 1.23 |
ENST00000604621.1 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
| chr19_-_51456321 | 1.16 |
ENST00000391809.2 |
KLK5 |
kallikrein-related peptidase 5 |
| chr19_-_51456344 | 1.16 |
ENST00000336334.3 ENST00000593428.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr19_-_51456198 | 1.03 |
ENST00000594846.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr16_+_68771128 | 0.88 |
ENST00000261769.5 ENST00000422392.2 |
CDH1 |
cadherin 1, type 1, E-cadherin (epithelial) |
| chr19_-_6720686 | 0.82 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr20_+_58179582 | 0.75 |
ENST00000371015.1 ENST00000395639.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr14_-_105635090 | 0.72 |
ENST00000331782.3 ENST00000347004.2 |
JAG2 |
jagged 2 |
| chr17_-_39677971 | 0.68 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr6_-_136847610 | 0.64 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr17_+_54671047 | 0.54 |
ENST00000332822.4 |
NOG |
noggin |
| chr8_-_75233563 | 0.49 |
ENST00000342232.4 |
JPH1 |
junctophilin 1 |
| chr1_-_153363452 | 0.49 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr8_-_142011036 | 0.46 |
ENST00000520892.1 |
PTK2 |
protein tyrosine kinase 2 |
| chrX_+_102585124 | 0.43 |
ENST00000332431.4 ENST00000372666.1 |
TCEAL7 |
transcription elongation factor A (SII)-like 7 |
| chr18_+_33877654 | 0.41 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr4_-_87374283 | 0.36 |
ENST00000361569.2 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr8_+_32405785 | 0.36 |
ENST00000287842.3 |
NRG1 |
neuregulin 1 |
| chr2_+_223289208 | 0.36 |
ENST00000321276.7 |
SGPP2 |
sphingosine-1-phosphate phosphatase 2 |
| chr6_+_7541845 | 0.35 |
ENST00000418664.2 |
DSP |
desmoplakin |
| chr6_+_7541808 | 0.34 |
ENST00000379802.3 |
DSP |
desmoplakin |
| chr8_-_22550815 | 0.32 |
ENST00000317216.2 |
EGR3 |
early growth response 3 |
| chr2_+_106468204 | 0.29 |
ENST00000425756.1 ENST00000393349.2 |
NCK2 |
NCK adaptor protein 2 |
| chr12_+_54447637 | 0.29 |
ENST00000609810.1 ENST00000430889.2 |
HOXC4 HOXC4 |
homeobox C4 Homeobox protein Hox-C4 |
| chr8_-_66754172 | 0.29 |
ENST00000401827.3 |
PDE7A |
phosphodiesterase 7A |
| chr6_+_112375275 | 0.28 |
ENST00000368666.2 ENST00000604763.1 ENST00000230529.5 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
| chr19_+_34287751 | 0.26 |
ENST00000590771.1 ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr6_-_13486369 | 0.25 |
ENST00000558378.1 |
AL583828.1 |
AL583828.1 |
| chr17_+_7788104 | 0.25 |
ENST00000380358.4 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
| chr8_-_144655141 | 0.24 |
ENST00000398882.3 |
MROH6 |
maestro heat-like repeat family member 6 |
| chr8_+_32406179 | 0.23 |
ENST00000405005.3 |
NRG1 |
neuregulin 1 |
| chr12_-_12715266 | 0.23 |
ENST00000228862.2 |
DUSP16 |
dual specificity phosphatase 16 |
| chr1_+_212782012 | 0.22 |
ENST00000341491.4 ENST00000366985.1 |
ATF3 |
activating transcription factor 3 |
| chr2_+_234104079 | 0.22 |
ENST00000417661.1 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
| chr22_+_32750872 | 0.21 |
ENST00000397468.1 |
RFPL3 |
ret finger protein-like 3 |
| chr19_-_42759300 | 0.19 |
ENST00000222329.4 |
ERF |
Ets2 repressor factor |
| chr8_+_32406137 | 0.19 |
ENST00000521670.1 |
NRG1 |
neuregulin 1 |
| chrX_-_151903101 | 0.19 |
ENST00000393900.3 |
MAGEA12 |
melanoma antigen family A, 12 |
| chr4_+_74735102 | 0.19 |
ENST00000395761.3 |
CXCL1 |
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chrX_-_151903184 | 0.19 |
ENST00000357916.4 ENST00000393869.3 |
MAGEA12 |
melanoma antigen family A, 12 |
| chr3_-_116163830 | 0.18 |
ENST00000333617.4 |
LSAMP |
limbic system-associated membrane protein |
| chr12_-_95044309 | 0.18 |
ENST00000261226.4 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
| chr19_-_51471362 | 0.18 |
ENST00000376853.4 ENST00000424910.2 |
KLK6 |
kallikrein-related peptidase 6 |
| chr19_+_44085189 | 0.18 |
ENST00000562365.2 |
PINLYP |
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr19_-_51471381 | 0.17 |
ENST00000594641.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr17_-_6459802 | 0.17 |
ENST00000262483.8 |
PITPNM3 |
PITPNM family member 3 |
| chr17_-_6459768 | 0.17 |
ENST00000421306.3 |
PITPNM3 |
PITPNM family member 3 |
| chr19_-_44306590 | 0.17 |
ENST00000377950.3 |
LYPD5 |
LY6/PLAUR domain containing 5 |
| chr6_+_112375462 | 0.16 |
ENST00000361714.1 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
| chr19_+_48867652 | 0.16 |
ENST00000344846.2 |
SYNGR4 |
synaptogyrin 4 |
| chr12_+_26348246 | 0.16 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr12_+_26348429 | 0.16 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr7_-_24797032 | 0.16 |
ENST00000409970.1 ENST00000409775.3 |
DFNA5 |
deafness, autosomal dominant 5 |
| chrX_+_151081351 | 0.16 |
ENST00000276344.2 |
MAGEA4 |
melanoma antigen family A, 4 |
| chr17_-_39728303 | 0.15 |
ENST00000588431.1 ENST00000246662.4 |
KRT9 |
keratin 9 |
| chr13_-_76056250 | 0.15 |
ENST00000377636.3 ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4 |
TBC1 domain family, member 4 |
| chr17_+_1958388 | 0.15 |
ENST00000399849.3 |
HIC1 |
hypermethylated in cancer 1 |
| chr2_-_46769694 | 0.15 |
ENST00000522587.1 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr16_+_27325202 | 0.15 |
ENST00000395762.2 ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R |
interleukin 4 receptor |
| chr12_-_123849374 | 0.15 |
ENST00000602398.1 ENST00000602750.1 |
SBNO1 |
strawberry notch homolog 1 (Drosophila) |
| chr7_+_39125365 | 0.15 |
ENST00000559001.1 ENST00000464276.2 |
POU6F2 |
POU class 6 homeobox 2 |
| chr3_-_48471454 | 0.14 |
ENST00000296440.6 ENST00000448774.2 |
PLXNB1 |
plexin B1 |
| chr12_-_71314617 | 0.14 |
ENST00000283228.2 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr4_-_100485143 | 0.14 |
ENST00000394877.3 |
TRMT10A |
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr7_-_102257139 | 0.14 |
ENST00000521076.1 ENST00000462172.1 ENST00000522801.1 ENST00000449970.2 ENST00000262940.7 |
RASA4 |
RAS p21 protein activator 4 |
| chr1_+_18958008 | 0.14 |
ENST00000420770.2 ENST00000400661.3 |
PAX7 |
paired box 7 |
| chr1_-_155947951 | 0.14 |
ENST00000313695.7 ENST00000497907.1 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr11_+_128562372 | 0.13 |
ENST00000344954.6 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr21_+_44073916 | 0.13 |
ENST00000349112.3 ENST00000398224.3 |
PDE9A |
phosphodiesterase 9A |
| chr7_-_51384451 | 0.13 |
ENST00000441453.1 ENST00000265136.7 ENST00000395542.2 ENST00000395540.2 |
COBL |
cordon-bleu WH2 repeat protein |
| chr13_-_99852916 | 0.13 |
ENST00000426037.2 ENST00000445737.2 |
UBAC2-AS1 |
UBAC2 antisense RNA 1 |
| chr20_-_36793663 | 0.12 |
ENST00000536701.1 ENST00000536724.1 |
TGM2 |
transglutaminase 2 |
| chr21_+_44073860 | 0.12 |
ENST00000335512.4 ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A |
phosphodiesterase 9A |
| chr7_-_71801980 | 0.12 |
ENST00000329008.5 |
CALN1 |
calneuron 1 |
| chr9_+_84304628 | 0.12 |
ENST00000437181.1 |
RP11-154D17.1 |
RP11-154D17.1 |
| chr17_+_35849937 | 0.12 |
ENST00000394389.4 |
DUSP14 |
dual specificity phosphatase 14 |
| chr4_-_100484825 | 0.12 |
ENST00000273962.3 ENST00000514547.1 ENST00000455368.2 |
TRMT10A |
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr19_+_44084696 | 0.12 |
ENST00000562255.1 ENST00000569031.2 |
PINLYP |
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr17_-_66597530 | 0.12 |
ENST00000592554.1 |
FAM20A |
family with sequence similarity 20, member A |
| chr7_-_752074 | 0.12 |
ENST00000360274.4 |
PRKAR1B |
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr17_-_43502987 | 0.11 |
ENST00000376922.2 |
ARHGAP27 |
Rho GTPase activating protein 27 |
| chr10_-_121356518 | 0.11 |
ENST00000369092.4 |
TIAL1 |
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr6_-_31560729 | 0.11 |
ENST00000340027.5 ENST00000376073.4 ENST00000376072.3 |
NCR3 |
natural cytotoxicity triggering receptor 3 |
| chr12_+_54943134 | 0.11 |
ENST00000243052.3 |
PDE1B |
phosphodiesterase 1B, calmodulin-dependent |
| chr19_-_49140609 | 0.10 |
ENST00000601104.1 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
| chr2_+_171571827 | 0.10 |
ENST00000375281.3 |
SP5 |
Sp5 transcription factor |
| chr11_+_46402744 | 0.10 |
ENST00000533952.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr11_+_2323236 | 0.10 |
ENST00000182290.4 |
TSPAN32 |
tetraspanin 32 |
| chr17_+_80376194 | 0.10 |
ENST00000337014.6 |
HEXDC |
hexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing |
| chr5_+_175299743 | 0.10 |
ENST00000502265.1 |
CPLX2 |
complexin 2 |
| chr19_+_6135646 | 0.10 |
ENST00000588304.1 ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2 |
acyl-CoA synthetase bubblegum family member 2 |
| chr9_+_132427883 | 0.10 |
ENST00000372469.4 |
PRRX2 |
paired related homeobox 2 |
| chr2_+_85661918 | 0.10 |
ENST00000340326.2 |
SH2D6 |
SH2 domain containing 6 |
| chr11_-_75062829 | 0.10 |
ENST00000393505.4 |
ARRB1 |
arrestin, beta 1 |
| chr1_-_156470556 | 0.09 |
ENST00000489057.1 ENST00000348159.4 |
MEF2D |
myocyte enhancer factor 2D |
| chr19_+_41305330 | 0.09 |
ENST00000593972.1 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
| chr8_-_145331153 | 0.09 |
ENST00000377412.4 |
KM-PA-2 |
KM-PA-2 protein; Uncharacterized protein |
| chr17_-_41132088 | 0.09 |
ENST00000591916.1 ENST00000451885.2 ENST00000454303.1 |
PTGES3L PTGES3L-AARSD1 |
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr3_+_57094469 | 0.09 |
ENST00000334325.1 |
SPATA12 |
spermatogenesis associated 12 |
| chr11_+_66886717 | 0.09 |
ENST00000398645.2 |
KDM2A |
lysine (K)-specific demethylase 2A |
| chr19_-_39226045 | 0.09 |
ENST00000597987.1 ENST00000595177.1 |
CAPN12 |
calpain 12 |
| chr10_+_127661942 | 0.09 |
ENST00000417114.1 ENST00000445510.1 ENST00000368691.1 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
| chr11_-_57089671 | 0.09 |
ENST00000532437.1 |
TNKS1BP1 |
tankyrase 1 binding protein 1, 182kDa |
| chr1_-_153538292 | 0.08 |
ENST00000497140.1 ENST00000368708.3 |
S100A2 |
S100 calcium binding protein A2 |
| chr11_-_75062730 | 0.08 |
ENST00000420843.2 ENST00000360025.3 |
ARRB1 |
arrestin, beta 1 |
| chr1_-_156470515 | 0.08 |
ENST00000340875.5 ENST00000368240.2 ENST00000353795.3 |
MEF2D |
myocyte enhancer factor 2D |
| chr4_-_38666430 | 0.08 |
ENST00000436901.1 |
AC021860.1 |
Uncharacterized protein |
| chr18_-_3874271 | 0.08 |
ENST00000400149.3 ENST00000400155.1 ENST00000400150.3 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr2_-_136873735 | 0.08 |
ENST00000409817.1 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
| chr18_-_21242774 | 0.08 |
ENST00000322980.9 |
ANKRD29 |
ankyrin repeat domain 29 |
| chr12_+_132413739 | 0.08 |
ENST00000443358.2 |
PUS1 |
pseudouridylate synthase 1 |
| chr19_-_33555780 | 0.08 |
ENST00000254260.3 ENST00000400226.4 |
RHPN2 |
rhophilin, Rho GTPase binding protein 2 |
| chr10_-_103603568 | 0.08 |
ENST00000356640.2 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr12_-_57522813 | 0.08 |
ENST00000556155.1 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr2_-_32236002 | 0.08 |
ENST00000404530.1 |
MEMO1 |
mediator of cell motility 1 |
| chr17_-_39216344 | 0.07 |
ENST00000391418.2 |
KRTAP2-3 |
keratin associated protein 2-3 |
| chr14_+_23563952 | 0.07 |
ENST00000319074.4 |
C14orf119 |
chromosome 14 open reading frame 119 |
| chr5_+_78908388 | 0.07 |
ENST00000296783.3 |
PAPD4 |
PAP associated domain containing 4 |
| chr12_+_26348582 | 0.07 |
ENST00000535504.1 |
SSPN |
sarcospan |
| chr16_+_30935418 | 0.07 |
ENST00000338343.4 |
FBXL19 |
F-box and leucine-rich repeat protein 19 |
| chr19_+_2164126 | 0.07 |
ENST00000398665.3 |
DOT1L |
DOT1-like histone H3K79 methyltransferase |
| chr19_-_19051927 | 0.07 |
ENST00000600077.1 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr1_-_204380919 | 0.07 |
ENST00000367188.4 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
| chr17_+_58499066 | 0.07 |
ENST00000474834.1 |
C17orf64 |
chromosome 17 open reading frame 64 |
| chr8_+_145064233 | 0.07 |
ENST00000529301.1 ENST00000395068.4 |
GRINA |
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr10_-_126849068 | 0.07 |
ENST00000494626.2 ENST00000337195.5 |
CTBP2 |
C-terminal binding protein 2 |
| chr6_+_29407083 | 0.07 |
ENST00000444197.2 |
OR10C1 |
olfactory receptor, family 10, subfamily C, member 1 (gene/pseudogene) |
| chr15_+_77712993 | 0.07 |
ENST00000336216.4 ENST00000381714.3 ENST00000558651.1 |
HMG20A |
high mobility group 20A |
| chr19_+_41305612 | 0.07 |
ENST00000594380.1 ENST00000593397.1 ENST00000601733.1 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
| chr19_+_41305627 | 0.06 |
ENST00000593525.1 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
| chr2_+_228337079 | 0.06 |
ENST00000409315.1 ENST00000373671.3 ENST00000409171.1 |
AGFG1 |
ArfGAP with FG repeats 1 |
| chr17_-_73840415 | 0.06 |
ENST00000592386.1 ENST00000412096.2 ENST00000586147.1 |
UNC13D |
unc-13 homolog D (C. elegans) |
| chr17_+_6918354 | 0.06 |
ENST00000552775.1 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr10_-_112064665 | 0.06 |
ENST00000369603.5 |
SMNDC1 |
survival motor neuron domain containing 1 |
| chr11_+_46403303 | 0.06 |
ENST00000407067.1 ENST00000395565.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chrX_+_37545012 | 0.06 |
ENST00000378616.3 |
XK |
X-linked Kx blood group (McLeod syndrome) |
| chr5_-_35195338 | 0.06 |
ENST00000509839.1 |
PRLR |
prolactin receptor |
| chr22_-_37976082 | 0.06 |
ENST00000215886.4 |
LGALS2 |
lectin, galactoside-binding, soluble, 2 |
| chr15_-_43513187 | 0.06 |
ENST00000540029.1 ENST00000441366.2 |
EPB42 |
erythrocyte membrane protein band 4.2 |
| chr6_-_35480705 | 0.06 |
ENST00000229771.6 |
TULP1 |
tubby like protein 1 |
| chr8_-_66753682 | 0.06 |
ENST00000396642.3 |
PDE7A |
phosphodiesterase 7A |
| chr12_+_132413798 | 0.06 |
ENST00000440818.2 ENST00000542167.2 ENST00000538037.1 ENST00000456665.2 |
PUS1 |
pseudouridylate synthase 1 |
| chr17_+_58499844 | 0.06 |
ENST00000269127.4 |
C17orf64 |
chromosome 17 open reading frame 64 |
| chr5_+_78908233 | 0.06 |
ENST00000453514.1 ENST00000423041.2 ENST00000504233.1 ENST00000428308.2 |
PAPD4 |
PAP associated domain containing 4 |
| chr19_+_41305085 | 0.06 |
ENST00000303961.4 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
| chr19_-_51336443 | 0.06 |
ENST00000598673.1 |
KLK15 |
kallikrein-related peptidase 15 |
| chrX_-_48693955 | 0.06 |
ENST00000218230.5 |
PCSK1N |
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr19_-_19051993 | 0.06 |
ENST00000594794.1 ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr3_-_52869205 | 0.05 |
ENST00000446157.2 |
MUSTN1 |
musculoskeletal, embryonic nuclear protein 1 |
| chr11_+_46403194 | 0.05 |
ENST00000395569.4 ENST00000395566.4 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr6_-_35480640 | 0.05 |
ENST00000428978.1 ENST00000322263.4 |
TULP1 |
tubby like protein 1 |
| chr17_-_77005860 | 0.05 |
ENST00000591773.1 ENST00000588611.1 ENST00000586916.2 ENST00000592033.1 ENST00000588075.1 ENST00000302345.2 ENST00000591811.1 |
CANT1 |
calcium activated nucleotidase 1 |
| chr11_+_66406088 | 0.05 |
ENST00000310092.7 ENST00000396053.4 ENST00000408993.2 |
RBM4 |
RNA binding motif protein 4 |
| chr12_+_132413765 | 0.05 |
ENST00000376649.3 ENST00000322060.5 |
PUS1 |
pseudouridylate synthase 1 |
| chr2_+_192543153 | 0.05 |
ENST00000425611.2 |
NABP1 |
nucleic acid binding protein 1 |
| chr10_-_103603523 | 0.05 |
ENST00000370046.1 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr11_-_129872712 | 0.05 |
ENST00000358825.5 ENST00000360871.3 ENST00000528746.1 |
PRDM10 |
PR domain containing 10 |
| chr13_-_52027134 | 0.05 |
ENST00000311234.4 ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6 |
integrator complex subunit 6 |
| chr19_+_41305740 | 0.05 |
ENST00000596517.1 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
| chr4_+_164265035 | 0.05 |
ENST00000338566.3 |
NPY5R |
neuropeptide Y receptor Y5 |
| chr19_+_13056663 | 0.05 |
ENST00000541222.1 ENST00000316856.3 ENST00000586534.1 ENST00000592268.1 |
RAD23A |
RAD23 homolog A (S. cerevisiae) |
| chr10_+_134145735 | 0.05 |
ENST00000368613.4 |
LRRC27 |
leucine rich repeat containing 27 |
| chr3_-_55521323 | 0.05 |
ENST00000264634.4 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
| chr12_+_57916584 | 0.05 |
ENST00000546632.1 ENST00000549623.1 ENST00000431731.2 |
MBD6 |
methyl-CpG binding domain protein 6 |
| chr3_+_8775466 | 0.05 |
ENST00000343849.2 ENST00000397368.2 |
CAV3 |
caveolin 3 |
| chrX_-_148571884 | 0.05 |
ENST00000537071.1 |
IDS |
iduronate 2-sulfatase |
| chrX_+_151867214 | 0.05 |
ENST00000329342.5 ENST00000412733.1 ENST00000457643.1 |
MAGEA6 |
melanoma antigen family A, 6 |
| chr10_+_52751010 | 0.05 |
ENST00000373985.1 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr2_-_74570520 | 0.05 |
ENST00000394019.2 ENST00000346834.4 ENST00000359484.4 ENST00000423644.1 ENST00000377634.4 ENST00000436454.1 |
SLC4A5 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr6_+_33387830 | 0.05 |
ENST00000293748.5 |
SYNGAP1 |
synaptic Ras GTPase activating protein 1 |
| chr6_+_33388013 | 0.05 |
ENST00000449372.2 |
SYNGAP1 |
synaptic Ras GTPase activating protein 1 |
| chr13_+_24734880 | 0.05 |
ENST00000382095.4 |
SPATA13 |
spermatogenesis associated 13 |
| chr18_-_3874247 | 0.05 |
ENST00000581699.1 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr1_+_220267429 | 0.05 |
ENST00000366922.1 ENST00000302637.5 |
IARS2 |
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr19_-_18654293 | 0.05 |
ENST00000597547.1 ENST00000222308.4 ENST00000544835.3 ENST00000610101.1 ENST00000597960.3 ENST00000608443.1 |
FKBP8 |
FK506 binding protein 8, 38kDa |
| chr7_-_128045984 | 0.05 |
ENST00000470772.1 ENST00000480861.1 ENST00000496200.1 |
IMPDH1 |
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr15_+_21145765 | 0.05 |
ENST00000553416.1 |
CT60 |
cancer/testis antigen 60 (non-protein coding) |
| chr1_-_31712401 | 0.05 |
ENST00000373736.2 |
NKAIN1 |
Na+/K+ transporting ATPase interacting 1 |
| chr12_-_8088871 | 0.05 |
ENST00000075120.7 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr5_+_76145920 | 0.05 |
ENST00000317593.4 |
S100Z |
S100 calcium binding protein Z |
| chrX_-_8769378 | 0.05 |
ENST00000543214.1 ENST00000381003.3 |
FAM9A |
family with sequence similarity 9, member A |
| chr15_-_70390191 | 0.05 |
ENST00000559191.1 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr18_-_3874752 | 0.05 |
ENST00000534970.1 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr5_+_76145826 | 0.05 |
ENST00000513010.1 |
S100Z |
S100 calcium binding protein Z |
| chr12_+_62654155 | 0.05 |
ENST00000312635.6 ENST00000393654.3 ENST00000549237.1 |
USP15 |
ubiquitin specific peptidase 15 |
| chr9_+_96026230 | 0.05 |
ENST00000448251.1 |
WNK2 |
WNK lysine deficient protein kinase 2 |
| chr12_-_16759711 | 0.05 |
ENST00000447609.1 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr13_+_28813645 | 0.05 |
ENST00000282391.5 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chrX_+_149737046 | 0.04 |
ENST00000370396.2 ENST00000542741.1 ENST00000543350.1 ENST00000424519.1 ENST00000413012.2 |
MTM1 |
myotubularin 1 |
| chr12_+_57916466 | 0.04 |
ENST00000355673.3 |
MBD6 |
methyl-CpG binding domain protein 6 |
| chr19_+_7828035 | 0.04 |
ENST00000327325.5 ENST00000394122.2 ENST00000248228.4 ENST00000334806.5 ENST00000359059.5 ENST00000357361.2 ENST00000596363.1 ENST00000595751.1 ENST00000596707.1 ENST00000597522.1 ENST00000595496.1 |
CLEC4M |
C-type lectin domain family 4, member M |
| chr9_-_34126730 | 0.04 |
ENST00000361264.4 |
DCAF12 |
DDB1 and CUL4 associated factor 12 |
| chr17_+_18761417 | 0.04 |
ENST00000419284.2 ENST00000268835.2 ENST00000412418.1 ENST00000575228.1 ENST00000575102.1 ENST00000536323.1 |
PRPSAP2 |
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr8_-_114389353 | 0.04 |
ENST00000343508.3 |
CSMD3 |
CUB and Sushi multiple domains 3 |
| chr9_-_21994344 | 0.04 |
ENST00000530628.2 ENST00000361570.3 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr19_+_41305406 | 0.04 |
ENST00000406058.2 ENST00000593726.1 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
| chr17_-_46703826 | 0.04 |
ENST00000550387.1 ENST00000311177.5 |
HOXB9 |
homeobox B9 |
| chr9_-_21994597 | 0.04 |
ENST00000579755.1 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr22_-_51066521 | 0.04 |
ENST00000395621.3 ENST00000395619.3 ENST00000356098.5 ENST00000216124.5 ENST00000453344.2 ENST00000547307.1 ENST00000547805.1 |
ARSA |
arylsulfatase A |
| chr5_-_142065223 | 0.04 |
ENST00000378046.1 |
FGF1 |
fibroblast growth factor 1 (acidic) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.3 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.3 | 0.8 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 1.2 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.1 | 0.8 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.9 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.1 | 1.0 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.2 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.2 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.0 | 0.3 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.5 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.1 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) response to odorant(GO:1990834) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.4 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.2 | GO:0090240 | follicle-stimulating hormone signaling pathway(GO:0042699) positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0061346 | cardiac right atrium morphogenesis(GO:0003213) negative regulation of melanin biosynthetic process(GO:0048022) positive regulation of anagen(GO:0051885) mediolateral intercalation(GO:0060031) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) negative regulation of secondary metabolite biosynthetic process(GO:1900377) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.5 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.0 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.3 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.2 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.1 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.2 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 4.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.6 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |