Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for RUNX3_BCL11A
Z-value: 0.81
Transcription factors associated with RUNX3_BCL11A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
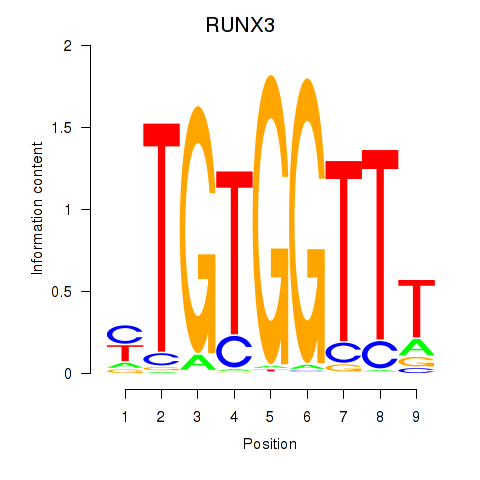
RUNX3
|
ENSG00000020633.14 | RUNX3 |
|
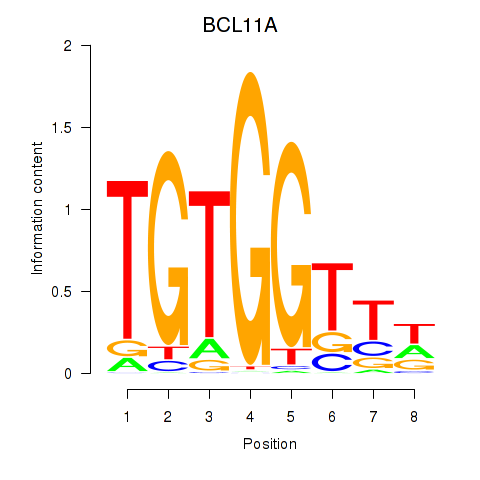
BCL11A
|
ENSG00000119866.16 | BCL11A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BCL11A | hg19_v2_chr2_-_60780536_60780541 | -0.97 | 8.3e-05 | Click! |
| RUNX3 | hg19_v2_chr1_-_25256368_25256476 | -0.75 | 3.4e-02 | Click! |
Activity profile of RUNX3_BCL11A motif
Sorted Z-values of RUNX3_BCL11A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RUNX3_BCL11A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_33359646 | 1.52 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359687 | 1.49 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr16_+_12995614 | 0.76 |
ENST00000423335.2 |
SHISA9 |
shisa family member 9 |
| chr2_-_158300556 | 0.58 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr21_-_36421626 | 0.54 |
ENST00000300305.3 |
RUNX1 |
runt-related transcription factor 1 |
| chr13_-_38172863 | 0.52 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr21_-_36421535 | 0.47 |
ENST00000416754.1 ENST00000437180.1 ENST00000455571.1 |
RUNX1 |
runt-related transcription factor 1 |
| chr1_+_221051699 | 0.45 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr1_+_6615241 | 0.44 |
ENST00000333172.6 ENST00000328191.4 ENST00000351136.3 |
TAS1R1 |
taste receptor, type 1, member 1 |
| chr8_-_60031762 | 0.41 |
ENST00000361421.1 |
TOX |
thymocyte selection-associated high mobility group box |
| chr6_-_169364429 | 0.40 |
ENST00000444586.1 |
RP3-495K2.3 |
RP3-495K2.3 |
| chr5_+_75699149 | 0.40 |
ENST00000379730.3 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr3_+_141105705 | 0.38 |
ENST00000513258.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr5_+_75699040 | 0.38 |
ENST00000274364.6 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr1_-_149889382 | 0.37 |
ENST00000369145.1 ENST00000369146.3 |
SV2A |
synaptic vesicle glycoprotein 2A |
| chr3_-_49170405 | 0.36 |
ENST00000305544.4 ENST00000494831.1 |
LAMB2 |
laminin, beta 2 (laminin S) |
| chr3_+_141105235 | 0.36 |
ENST00000503809.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr6_-_31514516 | 0.34 |
ENST00000303892.5 ENST00000483251.1 |
ATP6V1G2 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr4_-_57976544 | 0.32 |
ENST00000295666.4 ENST00000537922.1 |
IGFBP7 |
insulin-like growth factor binding protein 7 |
| chr10_+_17272608 | 0.32 |
ENST00000421459.2 |
VIM |
vimentin |
| chr3_+_141043050 | 0.32 |
ENST00000509842.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr1_-_113162040 | 0.31 |
ENST00000358039.4 ENST00000369668.2 |
ST7L |
suppression of tumorigenicity 7 like |
| chr3_-_150920979 | 0.30 |
ENST00000309180.5 ENST00000480322.1 |
GPR171 |
G protein-coupled receptor 171 |
| chr17_-_29648761 | 0.29 |
ENST00000247270.3 ENST00000462804.2 |
EVI2A |
ecotropic viral integration site 2A |
| chr5_-_111091948 | 0.29 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr1_-_113161730 | 0.28 |
ENST00000544629.1 ENST00000543570.1 ENST00000360743.4 ENST00000490067.1 ENST00000343210.7 ENST00000369666.1 |
ST7L |
suppression of tumorigenicity 7 like |
| chr15_-_93632421 | 0.28 |
ENST00000329082.7 |
RGMA |
repulsive guidance molecule family member a |
| chr1_+_164528866 | 0.28 |
ENST00000420696.2 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr15_-_37393406 | 0.28 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr1_+_145727681 | 0.27 |
ENST00000417171.1 ENST00000451928.2 |
PDZK1 |
PDZ domain containing 1 |
| chrX_-_106960285 | 0.26 |
ENST00000503515.1 ENST00000372397.2 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr2_+_136499287 | 0.26 |
ENST00000415164.1 |
UBXN4 |
UBX domain protein 4 |
| chr17_+_38599693 | 0.26 |
ENST00000542955.1 ENST00000269593.4 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
| chr15_-_37392703 | 0.26 |
ENST00000382766.2 ENST00000444725.1 |
MEIS2 |
Meis homeobox 2 |
| chr6_+_144904334 | 0.25 |
ENST00000367526.4 |
UTRN |
utrophin |
| chr12_-_4754339 | 0.25 |
ENST00000228850.1 |
AKAP3 |
A kinase (PRKA) anchor protein 3 |
| chr1_-_184006611 | 0.25 |
ENST00000546159.1 |
COLGALT2 |
collagen beta(1-O)galactosyltransferase 2 |
| chr14_+_64680854 | 0.23 |
ENST00000458046.2 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
| chr14_+_51955831 | 0.23 |
ENST00000356218.4 |
FRMD6 |
FERM domain containing 6 |
| chr9_+_75766763 | 0.22 |
ENST00000456643.1 ENST00000415424.1 |
ANXA1 |
annexin A1 |
| chr10_+_17270214 | 0.22 |
ENST00000544301.1 |
VIM |
vimentin |
| chr1_-_86043921 | 0.22 |
ENST00000535924.2 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr3_-_101395936 | 0.21 |
ENST00000461821.1 |
ZBTB11 |
zinc finger and BTB domain containing 11 |
| chr5_+_131993856 | 0.21 |
ENST00000304506.3 |
IL13 |
interleukin 13 |
| chr3_+_50126341 | 0.21 |
ENST00000347869.3 ENST00000469838.1 ENST00000404526.2 ENST00000441305.1 |
RBM5 |
RNA binding motif protein 5 |
| chr7_+_75028199 | 0.21 |
ENST00000437796.1 |
TRIM73 |
tripartite motif containing 73 |
| chr14_+_85996507 | 0.21 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr15_-_43910998 | 0.21 |
ENST00000450892.2 |
STRC |
stereocilin |
| chr9_-_130541017 | 0.21 |
ENST00000314830.8 |
SH2D3C |
SH2 domain containing 3C |
| chr9_-_123342415 | 0.20 |
ENST00000349780.4 ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chrX_+_22050546 | 0.20 |
ENST00000379374.4 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr2_-_175462934 | 0.20 |
ENST00000392546.2 ENST00000436221.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr19_-_17375541 | 0.19 |
ENST00000252597.3 |
USHBP1 |
Usher syndrome 1C binding protein 1 |
| chr16_+_2802623 | 0.19 |
ENST00000576924.1 ENST00000575009.1 ENST00000576415.1 ENST00000571378.1 |
SRRM2 |
serine/arginine repetitive matrix 2 |
| chr10_+_89420706 | 0.19 |
ENST00000427144.2 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr2_-_111291587 | 0.19 |
ENST00000437167.1 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
| chr11_-_102826434 | 0.18 |
ENST00000340273.4 ENST00000260302.3 |
MMP13 |
matrix metallopeptidase 13 (collagenase 3) |
| chr12_+_69139886 | 0.18 |
ENST00000398004.2 |
SLC35E3 |
solute carrier family 35, member E3 |
| chrX_+_48542168 | 0.18 |
ENST00000376701.4 |
WAS |
Wiskott-Aldrich syndrome |
| chr14_+_85996471 | 0.18 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr3_+_96533413 | 0.18 |
ENST00000470610.2 ENST00000389672.5 |
EPHA6 |
EPH receptor A6 |
| chr15_+_41913690 | 0.18 |
ENST00000563576.1 |
MGA |
MGA, MAX dimerization protein |
| chr2_+_228678550 | 0.18 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chr1_+_39876151 | 0.17 |
ENST00000530275.1 |
KIAA0754 |
KIAA0754 |
| chr20_+_43991668 | 0.17 |
ENST00000243918.5 ENST00000426004.1 |
SYS1 |
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr11_-_65548265 | 0.17 |
ENST00000532090.2 |
AP5B1 |
adaptor-related protein complex 5, beta 1 subunit |
| chr11_+_108093559 | 0.17 |
ENST00000278616.4 |
ATM |
ataxia telangiectasia mutated |
| chr3_+_98250743 | 0.17 |
ENST00000284311.3 |
GPR15 |
G protein-coupled receptor 15 |
| chr13_-_99910673 | 0.16 |
ENST00000397473.2 ENST00000397470.2 |
GPR18 |
G protein-coupled receptor 18 |
| chr11_-_2323290 | 0.16 |
ENST00000381153.3 |
C11orf21 |
chromosome 11 open reading frame 21 |
| chr12_-_53594227 | 0.16 |
ENST00000550743.2 |
ITGB7 |
integrin, beta 7 |
| chr9_-_20622478 | 0.16 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr12_+_75874984 | 0.16 |
ENST00000550491.1 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr4_+_71091786 | 0.16 |
ENST00000317987.5 |
FDCSP |
follicular dendritic cell secreted protein |
| chr22_+_40440804 | 0.16 |
ENST00000441751.1 ENST00000301923.9 |
TNRC6B |
trinucleotide repeat containing 6B |
| chr7_-_141646726 | 0.16 |
ENST00000438351.1 ENST00000439991.1 ENST00000551012.2 ENST00000546910.1 |
CLEC5A |
C-type lectin domain family 5, member A |
| chr6_-_75912508 | 0.15 |
ENST00000416123.2 |
COL12A1 |
collagen, type XII, alpha 1 |
| chr8_+_27183033 | 0.15 |
ENST00000420218.2 |
PTK2B |
protein tyrosine kinase 2 beta |
| chrX_-_40036520 | 0.15 |
ENST00000406200.2 ENST00000378455.4 ENST00000342274.4 |
BCOR |
BCL6 corepressor |
| chr1_+_17634689 | 0.15 |
ENST00000375453.1 ENST00000375448.4 |
PADI4 |
peptidyl arginine deiminase, type IV |
| chr3_+_49977440 | 0.15 |
ENST00000442092.1 ENST00000266022.4 ENST00000443081.1 |
RBM6 |
RNA binding motif protein 6 |
| chr8_+_27182862 | 0.15 |
ENST00000521164.1 ENST00000346049.5 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr18_+_7946941 | 0.15 |
ENST00000444013.1 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
| chrX_+_47441712 | 0.15 |
ENST00000218388.4 ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
| chr11_-_67141090 | 0.14 |
ENST00000312438.7 |
CLCF1 |
cardiotrophin-like cytokine factor 1 |
| chr13_+_28527647 | 0.14 |
ENST00000567234.1 |
LINC00543 |
long intergenic non-protein coding RNA 543 |
| chr7_+_120628731 | 0.14 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr1_-_235098861 | 0.14 |
ENST00000458044.1 |
RP11-443B7.1 |
RP11-443B7.1 |
| chr8_-_72274467 | 0.14 |
ENST00000340726.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr10_-_65028817 | 0.14 |
ENST00000542921.1 |
JMJD1C |
jumonji domain containing 1C |
| chr4_-_57524061 | 0.14 |
ENST00000508121.1 |
HOPX |
HOP homeobox |
| chr21_-_31744557 | 0.14 |
ENST00000399889.2 |
KRTAP13-2 |
keratin associated protein 13-2 |
| chr9_-_34048873 | 0.13 |
ENST00000449054.1 ENST00000379239.4 ENST00000539807.1 ENST00000379238.1 ENST00000418786.2 ENST00000360802.1 ENST00000412543.1 |
UBAP2 |
ubiquitin associated protein 2 |
| chr2_+_68872954 | 0.13 |
ENST00000394342.2 |
PROKR1 |
prokineticin receptor 1 |
| chr17_-_47287928 | 0.13 |
ENST00000507680.1 |
GNGT2 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr7_+_116660246 | 0.13 |
ENST00000434836.1 ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7 |
suppression of tumorigenicity 7 |
| chr1_+_12524965 | 0.13 |
ENST00000471923.1 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr10_-_14372870 | 0.13 |
ENST00000357447.2 |
FRMD4A |
FERM domain containing 4A |
| chr9_+_131799213 | 0.13 |
ENST00000358369.4 ENST00000406926.2 ENST00000277475.5 ENST00000450073.1 |
FAM73B |
family with sequence similarity 73, member B |
| chr12_-_112819896 | 0.13 |
ENST00000377560.5 ENST00000430131.2 ENST00000550722.1 ENST00000550724.1 |
HECTD4 |
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr3_+_49977490 | 0.13 |
ENST00000539992.1 |
RBM6 |
RNA binding motif protein 6 |
| chr17_-_1531635 | 0.13 |
ENST00000571650.1 |
SLC43A2 |
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr3_+_49977523 | 0.13 |
ENST00000422955.1 |
RBM6 |
RNA binding motif protein 6 |
| chr10_-_65028938 | 0.12 |
ENST00000402544.1 |
JMJD1C |
jumonji domain containing 1C |
| chr12_+_79439405 | 0.12 |
ENST00000552744.1 |
SYT1 |
synaptotagmin I |
| chr13_-_103053946 | 0.12 |
ENST00000376131.4 |
FGF14 |
fibroblast growth factor 14 |
| chr1_-_168513229 | 0.12 |
ENST00000367819.2 |
XCL2 |
chemokine (C motif) ligand 2 |
| chr17_-_56082455 | 0.12 |
ENST00000578794.1 |
RP11-159D12.5 |
Uncharacterized protein |
| chr9_-_117853297 | 0.12 |
ENST00000542877.1 ENST00000537320.1 ENST00000341037.4 |
TNC |
tenascin C |
| chr1_-_161039456 | 0.12 |
ENST00000368016.3 |
ARHGAP30 |
Rho GTPase activating protein 30 |
| chr6_+_30908747 | 0.12 |
ENST00000462446.1 ENST00000304311.2 |
DPCR1 |
diffuse panbronchiolitis critical region 1 |
| chr12_-_71031220 | 0.12 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr9_+_34652164 | 0.12 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chr8_+_27168988 | 0.11 |
ENST00000397501.1 ENST00000338238.4 ENST00000544172.1 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr10_-_13276329 | 0.11 |
ENST00000378681.3 ENST00000463405.2 |
UCMA |
upper zone of growth plate and cartilage matrix associated |
| chr20_+_43343886 | 0.11 |
ENST00000190983.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr22_+_23222886 | 0.11 |
ENST00000390319.2 |
IGLV3-1 |
immunoglobulin lambda variable 3-1 |
| chr20_-_45035198 | 0.11 |
ENST00000372176.1 |
ELMO2 |
engulfment and cell motility 2 |
| chr19_-_17375527 | 0.11 |
ENST00000431146.2 ENST00000594190.1 |
USHBP1 |
Usher syndrome 1C binding protein 1 |
| chr14_-_85996332 | 0.11 |
ENST00000380722.1 |
RP11-497E19.1 |
RP11-497E19.1 |
| chr1_+_110453109 | 0.11 |
ENST00000525659.1 |
CSF1 |
colony stimulating factor 1 (macrophage) |
| chr20_+_43343517 | 0.11 |
ENST00000372865.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr3_-_71179988 | 0.11 |
ENST00000491238.1 |
FOXP1 |
forkhead box P1 |
| chr16_+_31366455 | 0.11 |
ENST00000268296.4 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr19_-_14064114 | 0.11 |
ENST00000585607.1 ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1 |
podocan-like 1 |
| chr2_-_71221942 | 0.11 |
ENST00000272438.4 |
TEX261 |
testis expressed 261 |
| chr2_+_136499179 | 0.11 |
ENST00000272638.9 |
UBXN4 |
UBX domain protein 4 |
| chr1_+_117544366 | 0.11 |
ENST00000256652.4 ENST00000369470.1 |
CD101 |
CD101 molecule |
| chr3_-_71179699 | 0.10 |
ENST00000497355.1 |
FOXP1 |
forkhead box P1 |
| chr19_-_14048804 | 0.10 |
ENST00000254320.3 ENST00000586075.1 |
PODNL1 |
podocan-like 1 |
| chr2_-_55646957 | 0.10 |
ENST00000263630.8 |
CCDC88A |
coiled-coil domain containing 88A |
| chr20_+_43343476 | 0.10 |
ENST00000372868.2 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr2_-_175462456 | 0.10 |
ENST00000409891.1 ENST00000410117.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr19_-_14049184 | 0.10 |
ENST00000339560.5 |
PODNL1 |
podocan-like 1 |
| chr11_-_6426635 | 0.10 |
ENST00000608645.1 ENST00000608394.1 ENST00000529519.1 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr5_+_139505520 | 0.10 |
ENST00000333305.3 |
IGIP |
IgA-inducing protein |
| chr5_+_133451254 | 0.10 |
ENST00000517851.1 ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
| chr8_-_72274095 | 0.10 |
ENST00000303824.7 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr12_-_71031185 | 0.10 |
ENST00000548122.1 ENST00000551525.1 ENST00000550358.1 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr17_+_7239821 | 0.10 |
ENST00000158762.3 ENST00000570457.2 |
ACAP1 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr9_-_130517522 | 0.10 |
ENST00000373274.3 ENST00000420366.1 |
SH2D3C |
SH2 domain containing 3C |
| chr17_-_1532106 | 0.10 |
ENST00000301335.5 ENST00000382147.4 |
SLC43A2 |
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr20_+_31595406 | 0.10 |
ENST00000170150.3 |
BPIFB2 |
BPI fold containing family B, member 2 |
| chr2_-_55647057 | 0.10 |
ENST00000436346.1 |
CCDC88A |
coiled-coil domain containing 88A |
| chr8_-_116681221 | 0.10 |
ENST00000395715.3 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr13_-_28674693 | 0.10 |
ENST00000537084.1 ENST00000241453.7 ENST00000380982.4 |
FLT3 |
fms-related tyrosine kinase 3 |
| chr4_+_78783674 | 0.10 |
ENST00000315567.8 |
MRPL1 |
mitochondrial ribosomal protein L1 |
| chr1_+_111772435 | 0.10 |
ENST00000524472.1 |
CHI3L2 |
chitinase 3-like 2 |
| chr16_-_89785777 | 0.09 |
ENST00000561976.1 |
VPS9D1 |
VPS9 domain containing 1 |
| chr20_-_45035223 | 0.09 |
ENST00000450812.1 ENST00000290246.6 ENST00000439931.2 ENST00000396391.1 |
ELMO2 |
engulfment and cell motility 2 |
| chr11_+_75428857 | 0.09 |
ENST00000198801.5 |
MOGAT2 |
monoacylglycerol O-acyltransferase 2 |
| chr16_-_10652993 | 0.09 |
ENST00000536829.1 |
EMP2 |
epithelial membrane protein 2 |
| chr5_+_156607829 | 0.09 |
ENST00000422843.3 |
ITK |
IL2-inducible T-cell kinase |
| chr1_-_227505289 | 0.09 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr3_-_147124547 | 0.09 |
ENST00000491672.1 ENST00000383075.3 |
ZIC4 |
Zic family member 4 |
| chr2_-_220083076 | 0.09 |
ENST00000295750.4 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr6_+_27925019 | 0.09 |
ENST00000244623.1 |
OR2B6 |
olfactory receptor, family 2, subfamily B, member 6 |
| chr5_-_159546396 | 0.09 |
ENST00000523662.1 ENST00000456329.3 ENST00000307063.7 |
PWWP2A |
PWWP domain containing 2A |
| chr17_-_39183452 | 0.09 |
ENST00000361883.5 |
KRTAP1-5 |
keratin associated protein 1-5 |
| chr1_-_182641367 | 0.09 |
ENST00000508450.1 |
RGS8 |
regulator of G-protein signaling 8 |
| chr11_+_22696314 | 0.08 |
ENST00000532398.1 ENST00000433790.1 |
GAS2 |
growth arrest-specific 2 |
| chr9_+_118916082 | 0.08 |
ENST00000328252.3 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
| chr11_-_108093329 | 0.08 |
ENST00000278612.8 |
NPAT |
nuclear protein, ataxia-telangiectasia locus |
| chr11_-_5345582 | 0.08 |
ENST00000328813.2 |
OR51B2 |
olfactory receptor, family 51, subfamily B, member 2 |
| chr7_+_18535346 | 0.08 |
ENST00000405010.3 ENST00000406451.4 ENST00000428307.2 |
HDAC9 |
histone deacetylase 9 |
| chr5_-_127674883 | 0.08 |
ENST00000507835.1 |
FBN2 |
fibrillin 2 |
| chr6_+_74405501 | 0.08 |
ENST00000437994.2 ENST00000422508.2 |
CD109 |
CD109 molecule |
| chr6_+_86159821 | 0.08 |
ENST00000369651.3 |
NT5E |
5'-nucleotidase, ecto (CD73) |
| chr12_+_120884222 | 0.08 |
ENST00000551765.1 ENST00000229384.5 |
GATC |
glutamyl-tRNA(Gln) amidotransferase, subunit C |
| chr18_-_77276057 | 0.08 |
ENST00000597412.1 |
AC018445.1 |
Uncharacterized protein |
| chrX_-_118739835 | 0.08 |
ENST00000542113.1 ENST00000304449.5 |
NKRF |
NFKB repressing factor |
| chr1_-_179112173 | 0.08 |
ENST00000408940.3 ENST00000504405.1 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr6_+_167704838 | 0.08 |
ENST00000366829.2 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr19_-_44285401 | 0.08 |
ENST00000262888.3 |
KCNN4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr9_-_139642968 | 0.08 |
ENST00000341206.4 |
LCN6 |
lipocalin 6 |
| chr2_-_18741882 | 0.08 |
ENST00000381249.3 |
RDH14 |
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr17_-_4545170 | 0.08 |
ENST00000576394.1 ENST00000574640.1 |
ALOX15 |
arachidonate 15-lipoxygenase |
| chr2_-_217559517 | 0.08 |
ENST00000449583.1 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
| chr11_+_196738 | 0.08 |
ENST00000325113.4 ENST00000342593.5 |
ODF3 |
outer dense fiber of sperm tails 3 |
| chrX_-_19688475 | 0.08 |
ENST00000541422.1 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr1_+_168545711 | 0.08 |
ENST00000367818.3 |
XCL1 |
chemokine (C motif) ligand 1 |
| chr2_+_202316392 | 0.07 |
ENST00000194530.3 ENST00000392249.2 |
STRADB |
STE20-related kinase adaptor beta |
| chr1_-_92951607 | 0.07 |
ENST00000427103.1 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr3_+_40518599 | 0.07 |
ENST00000314686.5 ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619 |
zinc finger protein 619 |
| chr1_-_161039753 | 0.07 |
ENST00000368015.1 |
ARHGAP30 |
Rho GTPase activating protein 30 |
| chr16_-_2837949 | 0.07 |
ENST00000570702.1 |
PRSS33 |
protease, serine, 33 |
| chr12_-_8693469 | 0.07 |
ENST00000545274.1 ENST00000446457.2 |
CLEC4E |
C-type lectin domain family 4, member E |
| chr17_+_19281034 | 0.07 |
ENST00000308406.5 ENST00000299612.7 |
MAPK7 |
mitogen-activated protein kinase 7 |
| chr2_-_68052694 | 0.07 |
ENST00000457448.1 |
AC010987.6 |
AC010987.6 |
| chr14_-_35183886 | 0.07 |
ENST00000298159.6 |
CFL2 |
cofilin 2 (muscle) |
| chr19_-_44124019 | 0.07 |
ENST00000300811.3 |
ZNF428 |
zinc finger protein 428 |
| chr13_-_62001982 | 0.07 |
ENST00000409186.1 |
PCDH20 |
protocadherin 20 |
| chr8_-_19540266 | 0.07 |
ENST00000311540.4 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr11_-_19262486 | 0.07 |
ENST00000250024.4 |
E2F8 |
E2F transcription factor 8 |
| chr6_+_12290586 | 0.07 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr11_-_35287243 | 0.07 |
ENST00000464522.2 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr11_+_60995849 | 0.07 |
ENST00000537932.1 |
PGA4 |
pepsinogen 4, group I (pepsinogen A) |
| chr22_-_30642782 | 0.07 |
ENST00000249075.3 |
LIF |
leukemia inhibitory factor |
| chr1_-_161039647 | 0.07 |
ENST00000368013.3 |
ARHGAP30 |
Rho GTPase activating protein 30 |
| chr8_-_19540086 | 0.07 |
ENST00000332246.6 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.5 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.5 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.6 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.4 | GO:0072249 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.1 | 0.2 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.1 | 0.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.2 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.1 | 0.2 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.1 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.1 | 0.2 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.8 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.3 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.4 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.5 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.3 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.2 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:1903971 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.3 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0046086 | adenosine biosynthetic process(GO:0046086) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.3 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.2 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.2 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.1 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:1903517 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.0 | GO:1901656 | cellular response to mycotoxin(GO:0036146) glycoside transport(GO:1901656) cellular response to bile acid(GO:1903413) |
| 0.0 | 0.0 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.2 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.3 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.2 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.1 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 1.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.0 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 4.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |