Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
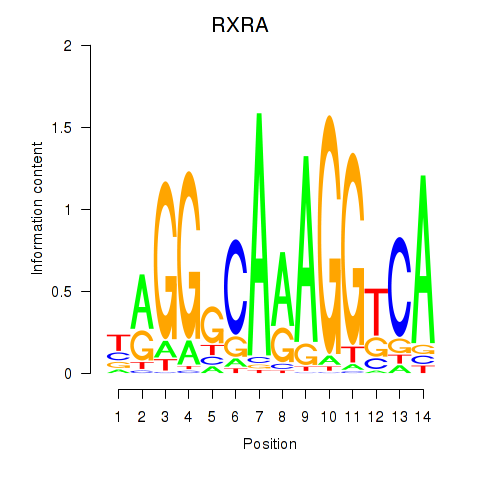
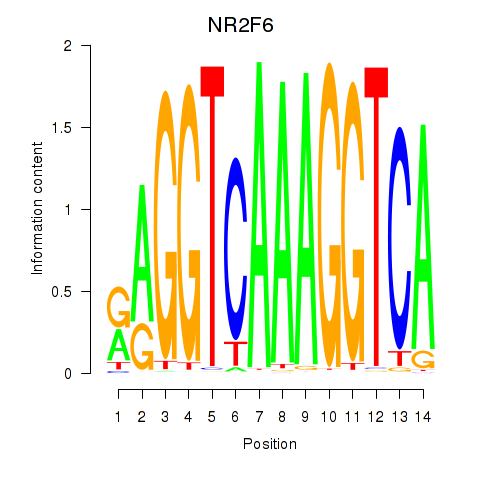
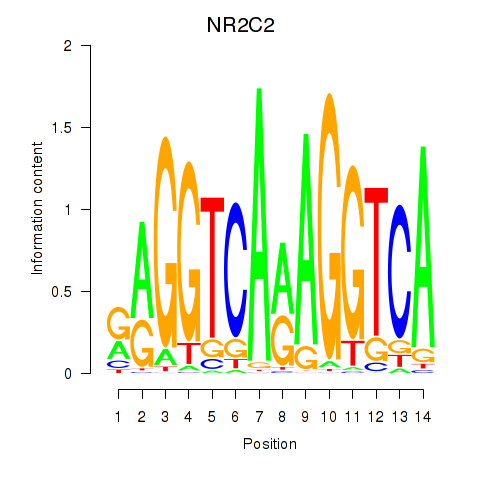
Results for RXRA_NR2F6_NR2C2
Z-value: 0.77
Transcription factors associated with RXRA_NR2F6_NR2C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRA
|
ENSG00000186350.8 | RXRA |
|
NR2F6
|
ENSG00000160113.5 | NR2F6 |
|
NR2C2
|
ENSG00000177463.11 | NR2C2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2C2 | hg19_v2_chr3_+_15045419_15045433 | 0.87 | 4.6e-03 | Click! |
| NR2F6 | hg19_v2_chr19_-_17356697_17356762 | 0.80 | 1.8e-02 | Click! |
| RXRA | hg19_v2_chr9_+_137298396_137298431, hg19_v2_chr9_+_137218362_137218426 | 0.68 | 6.1e-02 | Click! |
Activity profile of RXRA_NR2F6_NR2C2 motif
Sorted Z-values of RXRA_NR2F6_NR2C2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RXRA_NR2F6_NR2C2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_43885252 | 1.51 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr15_+_43985084 | 1.44 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr19_-_10687907 | 0.92 |
ENST00000589348.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr19_-_10687948 | 0.87 |
ENST00000592285.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr12_+_132413739 | 0.80 |
ENST00000443358.2 |
PUS1 |
pseudouridylate synthase 1 |
| chr19_-_10687983 | 0.77 |
ENST00000587069.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr1_+_65613340 | 0.76 |
ENST00000546702.1 |
AK4 |
adenylate kinase 4 |
| chr15_+_43985725 | 0.74 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr12_+_132413798 | 0.61 |
ENST00000440818.2 ENST00000542167.2 ENST00000538037.1 ENST00000456665.2 |
PUS1 |
pseudouridylate synthase 1 |
| chr12_+_132413765 | 0.58 |
ENST00000376649.3 ENST00000322060.5 |
PUS1 |
pseudouridylate synthase 1 |
| chr1_+_26869597 | 0.56 |
ENST00000530003.1 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr16_-_4401258 | 0.56 |
ENST00000577031.1 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr12_-_54779511 | 0.54 |
ENST00000551109.1 ENST00000546970.1 |
ZNF385A |
zinc finger protein 385A |
| chr7_+_86274145 | 0.52 |
ENST00000439827.1 ENST00000394720.2 ENST00000421579.1 |
GRM3 |
glutamate receptor, metabotropic 3 |
| chr7_+_86273952 | 0.50 |
ENST00000536043.1 |
GRM3 |
glutamate receptor, metabotropic 3 |
| chr16_-_4401284 | 0.50 |
ENST00000318059.3 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr13_-_103046837 | 0.43 |
ENST00000607251.1 |
FGF14-IT1 |
FGF14 intronic transcript 1 (non-protein coding) |
| chr19_+_39616410 | 0.42 |
ENST00000602004.1 ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4 |
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr9_-_139137648 | 0.40 |
ENST00000358701.5 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
| chr19_+_45394477 | 0.40 |
ENST00000252487.5 ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr19_-_6720686 | 0.40 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr7_+_86273700 | 0.40 |
ENST00000546348.1 |
GRM3 |
glutamate receptor, metabotropic 3 |
| chr4_-_7069760 | 0.39 |
ENST00000264954.4 |
GRPEL1 |
GrpE-like 1, mitochondrial (E. coli) |
| chr17_-_2614927 | 0.39 |
ENST00000435359.1 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
| chr20_-_43883197 | 0.39 |
ENST00000338380.2 |
SLPI |
secretory leukocyte peptidase inhibitor |
| chr3_-_119396193 | 0.37 |
ENST00000484810.1 ENST00000497116.1 ENST00000261070.2 |
COX17 |
COX17 cytochrome c oxidase copper chaperone |
| chr17_+_48609903 | 0.37 |
ENST00000268933.3 |
EPN3 |
epsin 3 |
| chr8_-_17555164 | 0.35 |
ENST00000297488.6 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr6_+_30882108 | 0.35 |
ENST00000541562.1 ENST00000421263.1 |
VARS2 |
valyl-tRNA synthetase 2, mitochondrial |
| chr1_+_65613513 | 0.35 |
ENST00000395334.2 |
AK4 |
adenylate kinase 4 |
| chr17_+_37894179 | 0.34 |
ENST00000577695.1 ENST00000309156.4 ENST00000309185.3 |
GRB7 |
growth factor receptor-bound protein 7 |
| chr1_+_65613852 | 0.33 |
ENST00000327299.7 |
AK4 |
adenylate kinase 4 |
| chr2_+_73441350 | 0.33 |
ENST00000389501.4 |
SMYD5 |
SMYD family member 5 |
| chr7_-_143105941 | 0.33 |
ENST00000275815.3 |
EPHA1 |
EPH receptor A1 |
| chr2_+_159313452 | 0.33 |
ENST00000389757.3 ENST00000389759.3 |
PKP4 |
plakophilin 4 |
| chr11_+_394196 | 0.32 |
ENST00000331563.2 ENST00000531857.1 |
PKP3 |
plakophilin 3 |
| chr17_+_1646130 | 0.31 |
ENST00000453066.1 ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr17_-_27503770 | 0.31 |
ENST00000533112.1 |
MYO18A |
myosin XVIIIA |
| chr17_+_48610074 | 0.31 |
ENST00000503690.1 ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3 |
epsin 3 |
| chr3_+_14989186 | 0.29 |
ENST00000435454.1 ENST00000323373.6 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr22_-_29784519 | 0.29 |
ENST00000357586.2 ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1 |
adaptor-related protein complex 1, beta 1 subunit |
| chr2_+_95940186 | 0.28 |
ENST00000403131.2 ENST00000317668.4 ENST00000317620.9 |
PROM2 |
prominin 2 |
| chr4_-_10023095 | 0.28 |
ENST00000264784.3 |
SLC2A9 |
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr19_+_1205740 | 0.27 |
ENST00000326873.7 |
STK11 |
serine/threonine kinase 11 |
| chr6_+_33172407 | 0.27 |
ENST00000374662.3 |
HSD17B8 |
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr11_-_123756334 | 0.27 |
ENST00000528595.1 ENST00000375026.2 |
TMEM225 |
transmembrane protein 225 |
| chr11_-_66496430 | 0.27 |
ENST00000533211.1 |
SPTBN2 |
spectrin, beta, non-erythrocytic 2 |
| chr6_+_125475335 | 0.26 |
ENST00000532429.1 ENST00000534199.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr2_-_85108164 | 0.25 |
ENST00000409520.2 |
TRABD2A |
TraB domain containing 2A |
| chr5_+_179220979 | 0.25 |
ENST00000292596.10 ENST00000401985.3 |
LTC4S |
leukotriene C4 synthase |
| chr9_+_74764278 | 0.24 |
ENST00000238018.4 ENST00000376989.3 |
GDA |
guanine deaminase |
| chr4_+_30721968 | 0.24 |
ENST00000361762.2 |
PCDH7 |
protocadherin 7 |
| chrX_-_153191708 | 0.23 |
ENST00000393721.1 ENST00000370028.3 |
ARHGAP4 |
Rho GTPase activating protein 4 |
| chr9_+_35673853 | 0.23 |
ENST00000378357.4 |
CA9 |
carbonic anhydrase IX |
| chr2_+_26568965 | 0.23 |
ENST00000260585.7 ENST00000447170.1 |
EPT1 |
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr6_+_43737939 | 0.21 |
ENST00000372067.3 |
VEGFA |
vascular endothelial growth factor A |
| chr13_+_32313658 | 0.21 |
ENST00000380314.1 ENST00000298386.2 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
| chrX_+_138612889 | 0.21 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr17_-_74497432 | 0.20 |
ENST00000590288.1 ENST00000313080.4 ENST00000592123.1 ENST00000591255.1 ENST00000585989.1 ENST00000591697.1 ENST00000389760.4 |
RHBDF2 |
rhomboid 5 homolog 2 (Drosophila) |
| chrX_-_117119243 | 0.19 |
ENST00000539496.1 ENST00000469946.1 |
KLHL13 |
kelch-like family member 13 |
| chr7_-_139763521 | 0.19 |
ENST00000263549.3 |
PARP12 |
poly (ADP-ribose) polymerase family, member 12 |
| chr19_-_17356697 | 0.19 |
ENST00000291442.3 |
NR2F6 |
nuclear receptor subfamily 2, group F, member 6 |
| chr8_+_28351707 | 0.19 |
ENST00000537916.1 ENST00000523546.1 ENST00000240093.3 |
FZD3 |
frizzled family receptor 3 |
| chr3_-_197300194 | 0.18 |
ENST00000358186.2 ENST00000431056.1 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
| chr1_-_16539094 | 0.18 |
ENST00000270747.3 |
ARHGEF19 |
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr19_-_42927251 | 0.18 |
ENST00000597001.1 |
LIPE |
lipase, hormone-sensitive |
| chr1_+_65613217 | 0.18 |
ENST00000545314.1 |
AK4 |
adenylate kinase 4 |
| chr19_-_48867291 | 0.18 |
ENST00000435956.3 |
TMEM143 |
transmembrane protein 143 |
| chrX_+_38420623 | 0.18 |
ENST00000378482.2 |
TSPAN7 |
tetraspanin 7 |
| chr19_-_46418033 | 0.18 |
ENST00000341294.2 |
NANOS2 |
nanos homolog 2 (Drosophila) |
| chr19_-_633576 | 0.18 |
ENST00000588649.2 |
POLRMT |
polymerase (RNA) mitochondrial (DNA directed) |
| chr2_+_95940220 | 0.18 |
ENST00000542147.1 |
PROM2 |
prominin 2 |
| chrX_-_153191674 | 0.18 |
ENST00000350060.5 ENST00000370016.1 |
ARHGAP4 |
Rho GTPase activating protein 4 |
| chr19_-_36304201 | 0.18 |
ENST00000301175.3 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr1_-_211848899 | 0.18 |
ENST00000366998.3 ENST00000540251.1 ENST00000366999.4 |
NEK2 |
NIMA-related kinase 2 |
| chr6_+_31926857 | 0.17 |
ENST00000375394.2 ENST00000544581.1 |
SKIV2L |
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr16_+_57673430 | 0.17 |
ENST00000540164.2 ENST00000568531.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr14_+_32546274 | 0.17 |
ENST00000396582.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr19_+_39390587 | 0.17 |
ENST00000572515.1 ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr1_+_154947126 | 0.17 |
ENST00000368439.1 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr1_+_154947148 | 0.17 |
ENST00000368436.1 ENST00000308987.5 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr17_-_39580775 | 0.17 |
ENST00000225550.3 |
KRT37 |
keratin 37 |
| chr16_+_57653989 | 0.17 |
ENST00000567835.1 ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr14_-_21490590 | 0.17 |
ENST00000557633.1 |
NDRG2 |
NDRG family member 2 |
| chr13_+_113344542 | 0.17 |
ENST00000487903.1 ENST00000375630.2 ENST00000375645.3 ENST00000283558.8 |
ATP11A |
ATPase, class VI, type 11A |
| chr16_+_57653854 | 0.16 |
ENST00000568908.1 ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr11_+_64073699 | 0.16 |
ENST00000405666.1 ENST00000468670.1 |
ESRRA |
estrogen-related receptor alpha |
| chr19_-_48867171 | 0.16 |
ENST00000377431.2 ENST00000436660.2 ENST00000541566.1 |
TMEM143 |
transmembrane protein 143 |
| chr7_-_72936608 | 0.15 |
ENST00000404251.1 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
| chr2_-_219134822 | 0.15 |
ENST00000444053.1 ENST00000248450.4 |
AAMP |
angio-associated, migratory cell protein |
| chr14_+_32546145 | 0.15 |
ENST00000556611.1 ENST00000539826.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr2_-_31637560 | 0.15 |
ENST00000379416.3 |
XDH |
xanthine dehydrogenase |
| chr8_-_80942061 | 0.15 |
ENST00000519386.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr8_-_145331153 | 0.15 |
ENST00000377412.4 |
KM-PA-2 |
KM-PA-2 protein; Uncharacterized protein |
| chr6_+_24775153 | 0.15 |
ENST00000356509.3 ENST00000230056.3 |
GMNN |
geminin, DNA replication inhibitor |
| chr19_+_49055332 | 0.14 |
ENST00000201586.2 |
SULT2B1 |
sulfotransferase family, cytosolic, 2B, member 1 |
| chr17_+_27369918 | 0.14 |
ENST00000323372.4 |
PIPOX |
pipecolic acid oxidase |
| chr1_-_156786530 | 0.14 |
ENST00000368198.3 |
SH2D2A |
SH2 domain containing 2A |
| chr1_-_156786634 | 0.14 |
ENST00000392306.2 ENST00000368199.3 |
SH2D2A |
SH2 domain containing 2A |
| chr12_-_51419924 | 0.14 |
ENST00000541174.2 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr2_+_219135115 | 0.14 |
ENST00000248451.3 ENST00000273077.4 |
PNKD |
paroxysmal nonkinesigenic dyskinesia |
| chr1_-_161102421 | 0.14 |
ENST00000490843.2 ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD |
death effector domain containing |
| chr19_-_35625765 | 0.13 |
ENST00000591633.1 |
LGI4 |
leucine-rich repeat LGI family, member 4 |
| chr15_-_82555000 | 0.13 |
ENST00000557844.1 ENST00000359445.3 ENST00000268206.7 |
EFTUD1 |
elongation factor Tu GTP binding domain containing 1 |
| chr6_+_143999072 | 0.13 |
ENST00000440869.2 ENST00000367582.3 ENST00000451827.2 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr12_+_130554803 | 0.13 |
ENST00000535487.1 |
RP11-474D1.2 |
RP11-474D1.2 |
| chr5_+_14664762 | 0.13 |
ENST00000284274.4 |
FAM105B |
family with sequence similarity 105, member B |
| chr2_-_207024134 | 0.13 |
ENST00000457011.1 ENST00000440274.1 ENST00000432169.1 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr12_-_51420108 | 0.13 |
ENST00000547198.1 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr4_+_95972822 | 0.13 |
ENST00000509540.1 ENST00000440890.2 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr14_+_105331596 | 0.13 |
ENST00000556508.1 ENST00000414716.3 ENST00000453495.1 ENST00000418279.1 |
CEP170B |
centrosomal protein 170B |
| chr19_-_51530916 | 0.13 |
ENST00000594768.1 |
KLK11 |
kallikrein-related peptidase 11 |
| chr10_-_103815874 | 0.13 |
ENST00000370033.4 ENST00000311122.5 |
C10orf76 |
chromosome 10 open reading frame 76 |
| chr2_-_75796837 | 0.13 |
ENST00000233712.1 |
EVA1A |
eva-1 homolog A (C. elegans) |
| chr2_+_159651821 | 0.13 |
ENST00000309950.3 ENST00000409042.1 |
DAPL1 |
death associated protein-like 1 |
| chr3_-_113465065 | 0.12 |
ENST00000497255.1 ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr14_-_24658053 | 0.12 |
ENST00000354464.6 |
IPO4 |
importin 4 |
| chr19_-_1174226 | 0.12 |
ENST00000587024.1 ENST00000361757.3 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
| chr22_-_20104700 | 0.12 |
ENST00000439169.2 ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A |
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr17_-_56350797 | 0.12 |
ENST00000577220.1 |
MPO |
myeloperoxidase |
| chr5_+_140345820 | 0.12 |
ENST00000289269.5 |
PCDHAC2 |
protocadherin alpha subfamily C, 2 |
| chr8_+_104831554 | 0.12 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr17_+_38333263 | 0.12 |
ENST00000456989.2 ENST00000543876.1 ENST00000544503.1 ENST00000264644.6 ENST00000538884.1 |
RAPGEFL1 |
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr6_+_106546808 | 0.12 |
ENST00000369089.3 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr8_-_80942139 | 0.12 |
ENST00000521434.1 ENST00000519120.1 ENST00000520946.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr2_+_74757050 | 0.12 |
ENST00000352222.3 ENST00000437202.1 |
HTRA2 |
HtrA serine peptidase 2 |
| chr3_+_57741957 | 0.11 |
ENST00000295951.3 |
SLMAP |
sarcolemma associated protein |
| chr8_+_124780672 | 0.11 |
ENST00000521166.1 ENST00000334705.7 |
FAM91A1 |
family with sequence similarity 91, member A1 |
| chr1_+_111888890 | 0.11 |
ENST00000369738.4 |
PIFO |
primary cilia formation |
| chr15_+_81591757 | 0.11 |
ENST00000558332.1 |
IL16 |
interleukin 16 |
| chr7_-_72936531 | 0.11 |
ENST00000339594.4 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
| chr22_-_51021397 | 0.11 |
ENST00000406938.2 |
CHKB |
choline kinase beta |
| chr8_+_145582633 | 0.11 |
ENST00000540505.1 |
SLC52A2 |
solute carrier family 52 (riboflavin transporter), member 2 |
| chr12_-_54778471 | 0.11 |
ENST00000550120.1 ENST00000394313.2 ENST00000547210.1 |
ZNF385A |
zinc finger protein 385A |
| chr14_-_106054659 | 0.11 |
ENST00000390539.2 |
IGHA2 |
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr19_+_14063278 | 0.11 |
ENST00000254337.6 |
DCAF15 |
DDB1 and CUL4 associated factor 15 |
| chr8_-_144815966 | 0.11 |
ENST00000388913.3 |
FAM83H |
family with sequence similarity 83, member H |
| chr22_+_29469012 | 0.11 |
ENST00000400335.4 ENST00000400338.2 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chrX_+_152912616 | 0.11 |
ENST00000342782.3 |
DUSP9 |
dual specificity phosphatase 9 |
| chr12_-_21757774 | 0.11 |
ENST00000261195.2 |
GYS2 |
glycogen synthase 2 (liver) |
| chr1_+_165864800 | 0.11 |
ENST00000469256.2 |
UCK2 |
uridine-cytidine kinase 2 |
| chr22_+_29469100 | 0.11 |
ENST00000327813.5 ENST00000407188.1 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chr1_-_248903150 | 0.11 |
ENST00000590317.1 |
LYPD8 |
LY6/PLAUR domain containing 8 |
| chr2_+_220325441 | 0.10 |
ENST00000396688.1 |
SPEG |
SPEG complex locus |
| chr3_-_48229846 | 0.10 |
ENST00000302506.3 ENST00000351231.3 ENST00000437972.1 |
CDC25A |
cell division cycle 25A |
| chr1_+_165864821 | 0.10 |
ENST00000470820.1 |
UCK2 |
uridine-cytidine kinase 2 |
| chr7_-_107204918 | 0.10 |
ENST00000297135.3 |
COG5 |
component of oligomeric golgi complex 5 |
| chr6_-_47010061 | 0.10 |
ENST00000371253.2 |
GPR110 |
G protein-coupled receptor 110 |
| chr6_-_146285221 | 0.10 |
ENST00000367503.3 ENST00000438092.2 ENST00000275233.7 |
SHPRH |
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr8_-_80942467 | 0.10 |
ENST00000518271.1 ENST00000276585.4 ENST00000521605.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr17_-_8027402 | 0.10 |
ENST00000541682.2 ENST00000317814.4 ENST00000577735.1 |
HES7 |
hes family bHLH transcription factor 7 |
| chr15_+_42120283 | 0.10 |
ENST00000542534.2 ENST00000397299.4 ENST00000408047.1 ENST00000431823.1 ENST00000382448.4 ENST00000342159.4 |
PLA2G4B JMJD7 JMJD7-PLA2G4B |
phospholipase A2, group IVB (cytosolic) jumonji domain containing 7 JMJD7-PLA2G4B readthrough |
| chr19_+_50145328 | 0.10 |
ENST00000360565.3 |
SCAF1 |
SR-related CTD-associated factor 1 |
| chr17_-_41132088 | 0.10 |
ENST00000591916.1 ENST00000451885.2 ENST00000454303.1 |
PTGES3L PTGES3L-AARSD1 |
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr17_-_73285293 | 0.10 |
ENST00000582778.1 ENST00000581988.1 ENST00000579207.1 ENST00000583332.1 ENST00000416858.2 ENST00000442286.2 ENST00000580151.1 ENST00000580994.1 ENST00000584438.1 ENST00000320362.3 ENST00000580273.1 |
SLC25A19 |
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr2_+_27719697 | 0.10 |
ENST00000264717.2 ENST00000424318.2 |
GCKR |
glucokinase (hexokinase 4) regulator |
| chr2_+_198365095 | 0.10 |
ENST00000409468.1 |
HSPE1 |
heat shock 10kDa protein 1 |
| chr4_+_123300591 | 0.10 |
ENST00000439307.1 ENST00000388724.2 |
ADAD1 |
adenosine deaminase domain containing 1 (testis-specific) |
| chr17_-_80017856 | 0.10 |
ENST00000577574.1 |
DUS1L |
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr1_+_12079517 | 0.10 |
ENST00000235332.4 ENST00000436478.2 |
MIIP |
migration and invasion inhibitory protein |
| chr18_-_11670159 | 0.10 |
ENST00000561598.1 |
RP11-677O4.2 |
RP11-677O4.2 |
| chr17_+_53828333 | 0.10 |
ENST00000268896.5 |
PCTP |
phosphatidylcholine transfer protein |
| chr6_-_146285455 | 0.10 |
ENST00000367505.2 |
SHPRH |
SNF2 histone linker PHD RING helicase, E3 ubiquitin protein ligase |
| chr1_+_33207381 | 0.10 |
ENST00000401073.2 |
KIAA1522 |
KIAA1522 |
| chr7_-_25702669 | 0.10 |
ENST00000446840.1 |
AC003090.1 |
AC003090.1 |
| chr16_+_2255710 | 0.10 |
ENST00000397124.1 ENST00000565250.1 |
MLST8 |
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr12_+_57916466 | 0.10 |
ENST00000355673.3 |
MBD6 |
methyl-CpG binding domain protein 6 |
| chrX_+_102585124 | 0.10 |
ENST00000332431.4 ENST00000372666.1 |
TCEAL7 |
transcription elongation factor A (SII)-like 7 |
| chr4_+_123300664 | 0.10 |
ENST00000388725.2 |
ADAD1 |
adenosine deaminase domain containing 1 (testis-specific) |
| chr6_+_159590423 | 0.09 |
ENST00000297267.9 ENST00000340366.6 |
FNDC1 |
fibronectin type III domain containing 1 |
| chr18_+_56530794 | 0.09 |
ENST00000590285.1 ENST00000586085.1 ENST00000589288.1 |
ZNF532 |
zinc finger protein 532 |
| chr16_+_30710462 | 0.09 |
ENST00000262518.4 ENST00000395059.2 ENST00000344771.4 |
SRCAP |
Snf2-related CREBBP activator protein |
| chr10_+_101542462 | 0.09 |
ENST00000370449.4 ENST00000370434.1 |
ABCC2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr11_+_46383121 | 0.09 |
ENST00000454345.1 |
DGKZ |
diacylglycerol kinase, zeta |
| chr17_+_53828381 | 0.09 |
ENST00000576183.1 |
PCTP |
phosphatidylcholine transfer protein |
| chr1_-_11107280 | 0.09 |
ENST00000400897.3 ENST00000400898.3 |
MASP2 |
mannan-binding lectin serine peptidase 2 |
| chr5_-_59995921 | 0.09 |
ENST00000453022.2 ENST00000545085.1 ENST00000265036.5 |
DEPDC1B |
DEP domain containing 1B |
| chr12_+_7079944 | 0.09 |
ENST00000261406.6 |
EMG1 |
EMG1 N1-specific pseudouridine methyltransferase |
| chr14_-_75643296 | 0.09 |
ENST00000303575.4 |
TMED10 |
transmembrane emp24-like trafficking protein 10 (yeast) |
| chr16_+_81812863 | 0.09 |
ENST00000359376.3 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| chr11_-_123525289 | 0.09 |
ENST00000392770.2 ENST00000299333.3 ENST00000530277.1 |
SCN3B |
sodium channel, voltage-gated, type III, beta subunit |
| chr7_-_74489609 | 0.09 |
ENST00000329959.4 ENST00000503250.2 ENST00000543840.1 |
WBSCR16 |
Williams-Beuren syndrome chromosome region 16 |
| chr19_+_17420340 | 0.09 |
ENST00000359866.4 |
DDA1 |
DET1 and DDB1 associated 1 |
| chrX_-_106362013 | 0.09 |
ENST00000372487.1 ENST00000372479.3 ENST00000203616.8 |
RBM41 |
RNA binding motif protein 41 |
| chr19_-_2328572 | 0.09 |
ENST00000252622.10 |
LSM7 |
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr5_+_176784837 | 0.09 |
ENST00000408923.3 |
RGS14 |
regulator of G-protein signaling 14 |
| chr14_-_77787198 | 0.09 |
ENST00000261534.4 |
POMT2 |
protein-O-mannosyltransferase 2 |
| chr6_+_149068464 | 0.09 |
ENST00000367463.4 |
UST |
uronyl-2-sulfotransferase |
| chr12_-_57522813 | 0.09 |
ENST00000556155.1 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr8_-_124286735 | 0.09 |
ENST00000395571.3 |
ZHX1 |
zinc fingers and homeoboxes 1 |
| chr16_+_4475806 | 0.09 |
ENST00000262375.6 ENST00000355296.4 ENST00000431375.2 ENST00000574895.1 |
DNAJA3 |
DnaJ (Hsp40) homolog, subfamily A, member 3 |
| chr22_-_37880543 | 0.09 |
ENST00000442496.1 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr22_+_38453378 | 0.09 |
ENST00000437453.1 ENST00000356976.3 |
PICK1 |
protein interacting with PRKCA 1 |
| chr3_-_185542817 | 0.09 |
ENST00000382199.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr12_+_49717081 | 0.09 |
ENST00000547807.1 ENST00000551567.1 |
TROAP |
trophinin associated protein |
| chrX_+_48916497 | 0.09 |
ENST00000496529.2 ENST00000376396.3 ENST00000422185.2 ENST00000603986.1 ENST00000536628.2 |
CCDC120 |
coiled-coil domain containing 120 |
| chr4_-_103266355 | 0.08 |
ENST00000424970.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr19_+_535835 | 0.08 |
ENST00000607527.1 ENST00000606065.1 |
CDC34 |
cell division cycle 34 |
| chr6_+_143772060 | 0.08 |
ENST00000367591.4 |
PEX3 |
peroxisomal biogenesis factor 3 |
| chr3_-_185542761 | 0.08 |
ENST00000457616.2 ENST00000346192.3 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.2 | 2.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 1.6 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 0.4 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.7 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.5 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.3 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 1.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 1.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.4 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.3 | GO:1904808 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.1 | 0.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.2 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.2 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.1 | 0.2 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 0.3 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.2 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 2.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.3 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0050787 | antibiotic metabolic process(GO:0016999) cellular chloride ion homeostasis(GO:0030644) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.1 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.3 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.0 | 0.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.4 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.3 | GO:0045059 | positive thymic T cell selection(GO:0045059) cellular response to UV-B(GO:0071493) |
| 0.0 | 0.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.0 | GO:0070873 | regulation of glycogen metabolic process(GO:0070873) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.1 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.3 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.1 | GO:0003069 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.0 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.5 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 0.3 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.3 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 2.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0071748 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 1.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.0 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0070069 | cytochrome complex(GO:0070069) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.3 | 1.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.3 | 2.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 1.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.3 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.3 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.3 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 2.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 3.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |