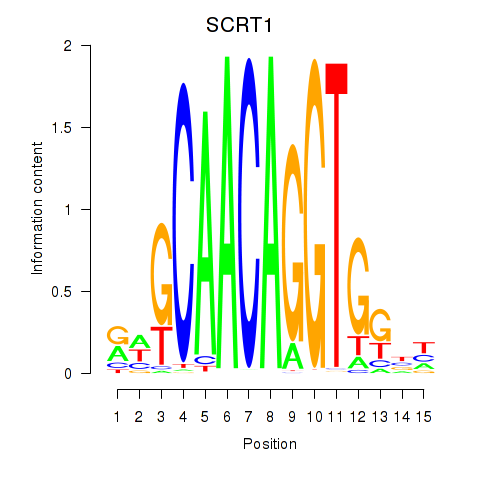
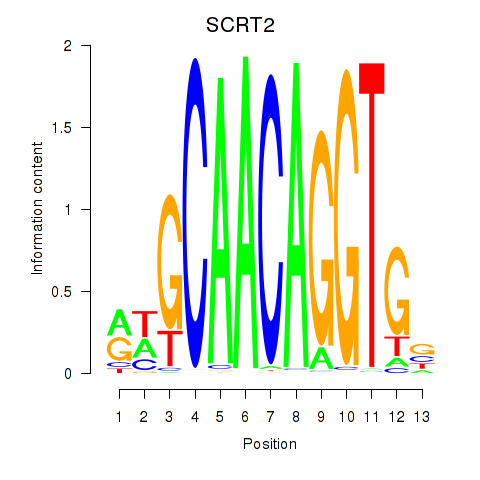
Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
Results for SCRT1_SCRT2
Z-value: 0.51
Transcription factors associated with SCRT1_SCRT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SCRT1
|
ENSG00000170616.9 | SCRT1 |
|
SCRT2
|
ENSG00000215397.3 | SCRT2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SCRT1 | hg19_v2_chr8_-_145559943_145559943 | -0.02 | 9.7e-01 | Click! |
Activity profile of SCRT1_SCRT2 motif
Sorted Z-values of SCRT1_SCRT2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SCRT1_SCRT2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_99986494 | 0.89 |
ENST00000372989.1 ENST00000455616.1 ENST00000454200.2 ENST00000276141.6 |
SYTL4 |
synaptotagmin-like 4 |
| chr6_-_31697255 | 0.82 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chrX_-_151143140 | 0.74 |
ENST00000393914.3 ENST00000370328.3 ENST00000370325.1 |
GABRE |
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr1_+_215256467 | 0.72 |
ENST00000391894.2 ENST00000444842.2 |
KCNK2 |
potassium channel, subfamily K, member 2 |
| chr4_-_6675550 | 0.63 |
ENST00000513179.1 ENST00000515205.1 |
RP11-539L10.3 |
RP11-539L10.3 |
| chr6_-_31697563 | 0.56 |
ENST00000375789.2 ENST00000416410.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr4_+_41258786 | 0.56 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr3_+_32280159 | 0.52 |
ENST00000458535.2 ENST00000307526.3 |
CMTM8 |
CKLF-like MARVEL transmembrane domain containing 8 |
| chr6_+_136172820 | 0.51 |
ENST00000308191.6 |
PDE7B |
phosphodiesterase 7B |
| chr5_+_15500280 | 0.49 |
ENST00000504595.1 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
| chr7_-_122526799 | 0.47 |
ENST00000334010.7 ENST00000313070.7 |
CADPS2 |
Ca++-dependent secretion activator 2 |
| chr17_+_78194205 | 0.47 |
ENST00000573809.1 ENST00000361193.3 ENST00000574967.1 ENST00000576126.1 ENST00000411502.3 ENST00000546047.2 |
SLC26A11 |
solute carrier family 26 (anion exchanger), member 11 |
| chrX_+_55744228 | 0.40 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr16_+_1031762 | 0.39 |
ENST00000293894.3 |
SOX8 |
SRY (sex determining region Y)-box 8 |
| chr19_+_12862604 | 0.37 |
ENST00000553030.1 |
BEST2 |
bestrophin 2 |
| chr2_-_175870085 | 0.36 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr3_-_114343768 | 0.36 |
ENST00000393785.2 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr4_-_90757364 | 0.36 |
ENST00000508895.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr12_-_33049690 | 0.35 |
ENST00000070846.6 ENST00000340811.4 |
PKP2 |
plakophilin 2 |
| chr5_+_32710736 | 0.35 |
ENST00000415685.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr2_+_101437487 | 0.34 |
ENST00000427413.1 ENST00000542504.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr17_-_15496722 | 0.31 |
ENST00000472534.1 |
CDRT1 |
CMT1A duplicated region transcript 1 |
| chr6_-_27440460 | 0.31 |
ENST00000377419.1 |
ZNF184 |
zinc finger protein 184 |
| chr6_-_27440837 | 0.31 |
ENST00000211936.6 |
ZNF184 |
zinc finger protein 184 |
| chr1_-_104238912 | 0.31 |
ENST00000330330.5 |
AMY1B |
amylase, alpha 1B (salivary) |
| chr1_-_104239076 | 0.31 |
ENST00000370080.3 |
AMY1B |
amylase, alpha 1B (salivary) |
| chr4_-_5890145 | 0.31 |
ENST00000397890.2 |
CRMP1 |
collapsin response mediator protein 1 |
| chr16_-_31161380 | 0.31 |
ENST00000569305.1 ENST00000418068.2 ENST00000268281.4 |
PRSS36 |
protease, serine, 36 |
| chr11_+_65779283 | 0.29 |
ENST00000312134.2 |
CST6 |
cystatin E/M |
| chrX_+_51927919 | 0.26 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chrX_+_51636629 | 0.26 |
ENST00000375722.1 ENST00000326587.7 ENST00000375695.2 |
MAGED1 |
melanoma antigen family D, 1 |
| chr1_+_150245099 | 0.25 |
ENST00000369099.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr2_-_224467093 | 0.25 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr16_-_29757272 | 0.25 |
ENST00000329410.3 |
C16orf54 |
chromosome 16 open reading frame 54 |
| chr12_-_54071181 | 0.24 |
ENST00000338662.5 |
ATP5G2 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chrX_-_51812268 | 0.24 |
ENST00000486010.1 ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B |
melanoma antigen family D, 4B |
| chr13_+_96743093 | 0.24 |
ENST00000376705.2 |
HS6ST3 |
heparan sulfate 6-O-sulfotransferase 3 |
| chr3_-_186080012 | 0.24 |
ENST00000544847.1 ENST00000265022.3 |
DGKG |
diacylglycerol kinase, gamma 90kDa |
| chr7_+_99699280 | 0.24 |
ENST00000421755.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr11_-_111794446 | 0.23 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr5_-_111091948 | 0.23 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chrX_+_51928002 | 0.23 |
ENST00000375626.3 |
MAGED4 |
melanoma antigen family D, 4 |
| chr4_-_89619386 | 0.22 |
ENST00000323061.5 |
NAP1L5 |
nucleosome assembly protein 1-like 5 |
| chr7_+_99699179 | 0.22 |
ENST00000438383.1 ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr13_+_76362974 | 0.21 |
ENST00000497947.2 |
LMO7 |
LIM domain 7 |
| chr6_+_108977520 | 0.20 |
ENST00000540898.1 |
FOXO3 |
forkhead box O3 |
| chr8_+_75736761 | 0.20 |
ENST00000260113.2 |
PI15 |
peptidase inhibitor 15 |
| chr22_-_22090043 | 0.20 |
ENST00000403503.1 |
YPEL1 |
yippee-like 1 (Drosophila) |
| chr17_+_7461613 | 0.19 |
ENST00000438470.1 ENST00000436057.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr19_-_18391708 | 0.19 |
ENST00000600972.1 |
JUND |
jun D proto-oncogene |
| chr20_-_45061695 | 0.19 |
ENST00000445496.2 |
ELMO2 |
engulfment and cell motility 2 |
| chr10_+_35484793 | 0.19 |
ENST00000488741.1 ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM |
cAMP responsive element modulator |
| chr7_+_12727250 | 0.19 |
ENST00000404894.1 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr3_+_141144963 | 0.19 |
ENST00000510726.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chrX_-_134232630 | 0.18 |
ENST00000535837.1 ENST00000433425.2 |
LINC00087 |
long intergenic non-protein coding RNA 87 |
| chr18_+_52495426 | 0.18 |
ENST00000262094.5 |
RAB27B |
RAB27B, member RAS oncogene family |
| chr19_-_11373128 | 0.18 |
ENST00000294618.7 |
DOCK6 |
dedicator of cytokinesis 6 |
| chr3_-_147124547 | 0.18 |
ENST00000491672.1 ENST00000383075.3 |
ZIC4 |
Zic family member 4 |
| chr19_-_46974664 | 0.18 |
ENST00000438932.2 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr19_-_46974741 | 0.18 |
ENST00000313683.10 ENST00000602246.1 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr14_-_77495007 | 0.18 |
ENST00000238647.3 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
| chr1_-_222885770 | 0.17 |
ENST00000355727.2 ENST00000340020.6 |
AIDA |
axin interactor, dorsalization associated |
| chr3_-_146187088 | 0.17 |
ENST00000497985.1 |
PLSCR2 |
phospholipid scramblase 2 |
| chr16_+_2039946 | 0.17 |
ENST00000248121.2 ENST00000568896.1 |
SYNGR3 |
synaptogyrin 3 |
| chr8_-_91657740 | 0.16 |
ENST00000422900.1 |
TMEM64 |
transmembrane protein 64 |
| chr8_-_116681221 | 0.16 |
ENST00000395715.3 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr11_+_61583721 | 0.15 |
ENST00000257261.6 |
FADS2 |
fatty acid desaturase 2 |
| chr4_-_120133661 | 0.15 |
ENST00000503243.1 ENST00000326780.3 |
RP11-455G16.1 |
Uncharacterized protein |
| chr3_+_16216137 | 0.15 |
ENST00000339732.5 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr1_+_104198377 | 0.15 |
ENST00000370083.4 |
AMY1A |
amylase, alpha 1A (salivary) |
| chr2_-_167232484 | 0.15 |
ENST00000375387.4 ENST00000303354.6 ENST00000409672.1 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
| chr16_+_2198604 | 0.15 |
ENST00000210187.6 |
RAB26 |
RAB26, member RAS oncogene family |
| chr16_+_67063262 | 0.14 |
ENST00000565389.1 |
CBFB |
core-binding factor, beta subunit |
| chr17_-_32484313 | 0.14 |
ENST00000359872.6 |
ASIC2 |
acid-sensing (proton-gated) ion channel 2 |
| chr12_+_103981044 | 0.14 |
ENST00000388887.2 |
STAB2 |
stabilin 2 |
| chr6_-_83903600 | 0.14 |
ENST00000506587.1 ENST00000507554.1 |
PGM3 |
phosphoglucomutase 3 |
| chr3_-_49851313 | 0.14 |
ENST00000333486.3 |
UBA7 |
ubiquitin-like modifier activating enzyme 7 |
| chr10_+_90672113 | 0.14 |
ENST00000371922.1 |
STAMBPL1 |
STAM binding protein-like 1 |
| chr10_-_47222824 | 0.14 |
ENST00000355232.3 |
AGAP10 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr1_-_201391149 | 0.13 |
ENST00000555948.1 ENST00000556362.1 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr6_+_121756809 | 0.13 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr17_+_39261584 | 0.12 |
ENST00000391415.1 |
KRTAP4-9 |
keratin associated protein 4-9 |
| chr3_-_72496035 | 0.12 |
ENST00000477973.2 |
RYBP |
RING1 and YY1 binding protein |
| chr10_+_35484053 | 0.12 |
ENST00000487763.1 ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM |
cAMP responsive element modulator |
| chr5_-_59064458 | 0.12 |
ENST00000502575.1 ENST00000507116.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr2_+_223162866 | 0.12 |
ENST00000295226.1 |
CCDC140 |
coiled-coil domain containing 140 |
| chr3_+_151986709 | 0.12 |
ENST00000495875.2 ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr8_-_91657909 | 0.12 |
ENST00000418210.2 |
TMEM64 |
transmembrane protein 64 |
| chr11_+_64002292 | 0.12 |
ENST00000426086.2 |
VEGFB |
vascular endothelial growth factor B |
| chr8_+_22429205 | 0.12 |
ENST00000520207.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr9_+_74526532 | 0.12 |
ENST00000486911.2 |
C9orf85 |
chromosome 9 open reading frame 85 |
| chr10_-_28623368 | 0.12 |
ENST00000441595.2 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr3_-_46904918 | 0.11 |
ENST00000395869.1 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr12_-_117318788 | 0.11 |
ENST00000550505.1 |
HRK |
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr17_-_1083078 | 0.11 |
ENST00000574266.1 ENST00000302538.5 |
ABR |
active BCR-related |
| chr21_-_35987438 | 0.11 |
ENST00000313806.4 |
RCAN1 |
regulator of calcineurin 1 |
| chr7_+_130126165 | 0.11 |
ENST00000427521.1 ENST00000416162.2 ENST00000378576.4 |
MEST |
mesoderm specific transcript |
| chr5_-_177423243 | 0.10 |
ENST00000308304.2 |
PROP1 |
PROP paired-like homeobox 1 |
| chr11_-_26593677 | 0.10 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr7_+_130126012 | 0.10 |
ENST00000341441.5 |
MEST |
mesoderm specific transcript |
| chr3_+_49044798 | 0.10 |
ENST00000438660.1 ENST00000608424.1 ENST00000415265.2 |
WDR6 |
WD repeat domain 6 |
| chr7_+_120628731 | 0.10 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr20_+_57427765 | 0.10 |
ENST00000371100.4 |
GNAS |
GNAS complex locus |
| chr17_+_63133587 | 0.10 |
ENST00000449996.3 ENST00000262406.9 |
RGS9 |
regulator of G-protein signaling 9 |
| chr1_-_9129895 | 0.10 |
ENST00000473209.1 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr17_-_33760164 | 0.10 |
ENST00000445092.1 ENST00000394562.1 ENST00000447040.2 |
SLFN12 |
schlafen family member 12 |
| chr3_-_46904946 | 0.09 |
ENST00000292327.4 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr17_-_33760269 | 0.09 |
ENST00000452764.3 |
SLFN12 |
schlafen family member 12 |
| chr2_-_24583314 | 0.09 |
ENST00000443927.1 ENST00000406921.3 ENST00000412011.1 |
ITSN2 |
intersectin 2 |
| chr1_-_95392635 | 0.09 |
ENST00000538964.1 ENST00000394202.4 ENST00000370206.4 |
CNN3 |
calponin 3, acidic |
| chr17_-_10101868 | 0.09 |
ENST00000432992.2 ENST00000540214.1 |
GAS7 |
growth arrest-specific 7 |
| chr19_-_7990991 | 0.09 |
ENST00000318978.4 |
CTXN1 |
cortexin 1 |
| chr4_-_140223670 | 0.09 |
ENST00000394228.1 ENST00000539387.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr12_+_117348742 | 0.09 |
ENST00000309909.5 ENST00000455858.2 |
FBXW8 |
F-box and WD repeat domain containing 8 |
| chr17_+_67410832 | 0.09 |
ENST00000590474.1 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
| chr1_+_43148625 | 0.09 |
ENST00000436427.1 |
YBX1 |
Y box binding protein 1 |
| chr22_+_42949925 | 0.09 |
ENST00000327678.5 ENST00000340239.4 ENST00000407614.4 ENST00000335879.5 |
SERHL2 |
serine hydrolase-like 2 |
| chr19_+_40877583 | 0.08 |
ENST00000596470.1 |
PLD3 |
phospholipase D family, member 3 |
| chr19_-_3062881 | 0.08 |
ENST00000586742.1 |
AES |
amino-terminal enhancer of split |
| chr3_+_141144954 | 0.08 |
ENST00000441582.2 ENST00000321464.5 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr5_-_73936451 | 0.08 |
ENST00000537006.1 |
ENC1 |
ectodermal-neural cortex 1 (with BTB domain) |
| chr8_-_133772794 | 0.08 |
ENST00000519187.1 ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71 |
transmembrane protein 71 |
| chr12_+_96337061 | 0.08 |
ENST00000266736.2 |
AMDHD1 |
amidohydrolase domain containing 1 |
| chr1_+_150522222 | 0.08 |
ENST00000369039.5 |
ADAMTSL4 |
ADAMTS-like 4 |
| chr14_+_102829300 | 0.08 |
ENST00000359520.7 |
TECPR2 |
tectonin beta-propeller repeat containing 2 |
| chr17_+_36858694 | 0.08 |
ENST00000563897.1 |
CTB-58E17.1 |
CTB-58E17.1 |
| chr2_-_24583583 | 0.08 |
ENST00000355123.4 |
ITSN2 |
intersectin 2 |
| chr1_-_24438664 | 0.07 |
ENST00000374434.3 ENST00000330966.7 ENST00000329601.7 |
MYOM3 |
myomesin 3 |
| chr16_-_1031259 | 0.07 |
ENST00000563837.1 ENST00000563863.1 ENST00000565069.1 ENST00000570014.1 |
RP11-161M6.2 LMF1 |
RP11-161M6.2 lipase maturation factor 1 |
| chr1_-_51425902 | 0.07 |
ENST00000396153.2 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
| chr1_+_33283043 | 0.07 |
ENST00000373476.1 ENST00000373475.5 ENST00000529027.1 ENST00000398243.3 |
S100PBP |
S100P binding protein |
| chr19_-_3063099 | 0.07 |
ENST00000221561.8 |
AES |
amino-terminal enhancer of split |
| chr6_-_45983549 | 0.07 |
ENST00000544153.1 |
CLIC5 |
chloride intracellular channel 5 |
| chr4_-_119757239 | 0.07 |
ENST00000280551.6 |
SEC24D |
SEC24 family member D |
| chr13_+_49551020 | 0.06 |
ENST00000541916.1 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr10_+_23216944 | 0.06 |
ENST00000298032.5 ENST00000409983.3 ENST00000409049.3 |
ARMC3 |
armadillo repeat containing 3 |
| chr1_-_154928562 | 0.06 |
ENST00000368463.3 ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1 |
pre-B-cell leukemia homeobox interacting protein 1 |
| chr9_+_114423615 | 0.06 |
ENST00000374293.4 |
GNG10 |
guanine nucleotide binding protein (G protein), gamma 10 |
| chr11_+_6624955 | 0.06 |
ENST00000299421.4 ENST00000537806.1 |
ILK |
integrin-linked kinase |
| chr4_-_52904425 | 0.06 |
ENST00000535450.1 |
SGCB |
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr14_-_64761078 | 0.06 |
ENST00000341099.4 ENST00000556275.1 ENST00000542956.1 ENST00000353772.3 ENST00000357782.2 ENST00000267525.6 |
ESR2 |
estrogen receptor 2 (ER beta) |
| chr11_-_26593649 | 0.06 |
ENST00000455601.2 |
MUC15 |
mucin 15, cell surface associated |
| chr10_+_23217006 | 0.06 |
ENST00000376528.4 ENST00000447081.1 |
ARMC3 |
armadillo repeat containing 3 |
| chr11_+_6624970 | 0.06 |
ENST00000420936.2 ENST00000528995.1 |
ILK |
integrin-linked kinase |
| chr4_-_110223523 | 0.06 |
ENST00000399127.1 |
COL25A1 |
collagen, type XXV, alpha 1 |
| chr11_-_30038490 | 0.06 |
ENST00000328224.6 |
KCNA4 |
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr9_-_116840728 | 0.05 |
ENST00000265132.3 |
AMBP |
alpha-1-microglobulin/bikunin precursor |
| chr2_-_165811756 | 0.05 |
ENST00000409662.1 |
SLC38A11 |
solute carrier family 38, member 11 |
| chr4_+_87856191 | 0.05 |
ENST00000503477.1 |
AFF1 |
AF4/FMR2 family, member 1 |
| chr5_+_138089100 | 0.05 |
ENST00000520339.1 ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr17_-_45056606 | 0.05 |
ENST00000322329.3 |
RPRML |
reprimo-like |
| chr11_-_117747607 | 0.05 |
ENST00000540359.1 ENST00000539526.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr1_+_156308403 | 0.05 |
ENST00000481479.1 ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC |
TSSK6 activating co-chaperone |
| chr4_-_141348789 | 0.05 |
ENST00000414773.1 |
CLGN |
calmegin |
| chr4_+_980785 | 0.05 |
ENST00000247933.4 ENST00000453894.1 |
IDUA |
iduronidase, alpha-L- |
| chr16_+_103816 | 0.05 |
ENST00000383018.3 ENST00000417493.1 |
SNRNP25 |
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr1_+_156308245 | 0.05 |
ENST00000368253.2 ENST00000470342.1 ENST00000368254.1 |
TSACC |
TSSK6 activating co-chaperone |
| chr4_-_119757322 | 0.04 |
ENST00000379735.5 |
SEC24D |
SEC24 family member D |
| chr16_+_30383613 | 0.04 |
ENST00000568749.1 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr12_-_55042140 | 0.04 |
ENST00000293371.6 ENST00000456047.2 |
DCD |
dermcidin |
| chr5_+_89854595 | 0.04 |
ENST00000405460.2 |
GPR98 |
G protein-coupled receptor 98 |
| chr11_-_117748138 | 0.04 |
ENST00000527717.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr3_-_42846021 | 0.04 |
ENST00000321331.7 |
HIGD1A |
HIG1 hypoxia inducible domain family, member 1A |
| chr11_-_26593779 | 0.04 |
ENST00000529533.1 |
MUC15 |
mucin 15, cell surface associated |
| chr11_-_67169253 | 0.04 |
ENST00000527663.1 ENST00000312989.7 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr3_-_42845951 | 0.04 |
ENST00000418900.2 ENST00000430190.1 |
HIGD1A |
HIG1 hypoxia inducible domain family, member 1A |
| chr16_+_67063855 | 0.04 |
ENST00000563939.2 |
CBFB |
core-binding factor, beta subunit |
| chr4_+_41540160 | 0.04 |
ENST00000503057.1 ENST00000511496.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr3_-_179754556 | 0.04 |
ENST00000263962.8 |
PEX5L |
peroxisomal biogenesis factor 5-like |
| chr2_-_241759622 | 0.04 |
ENST00000320389.7 ENST00000498729.2 |
KIF1A |
kinesin family member 1A |
| chrX_-_15332665 | 0.04 |
ENST00000537676.1 ENST00000344384.4 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr4_-_10117949 | 0.04 |
ENST00000508079.1 |
WDR1 |
WD repeat domain 1 |
| chr4_+_70146217 | 0.04 |
ENST00000335568.5 ENST00000511240.1 |
UGT2B28 |
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr9_+_99212403 | 0.03 |
ENST00000375251.3 ENST00000375249.4 |
HABP4 |
hyaluronan binding protein 4 |
| chrX_-_153979315 | 0.03 |
ENST00000369575.3 ENST00000369568.4 ENST00000424127.2 |
GAB3 |
GRB2-associated binding protein 3 |
| chr16_-_50402690 | 0.03 |
ENST00000394689.2 |
BRD7 |
bromodomain containing 7 |
| chr22_-_19466683 | 0.03 |
ENST00000399523.1 ENST00000421968.2 ENST00000447868.1 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
| chr1_-_9129631 | 0.03 |
ENST00000377414.3 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr22_-_22090064 | 0.03 |
ENST00000339468.3 |
YPEL1 |
yippee-like 1 (Drosophila) |
| chr7_+_138145076 | 0.03 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr2_+_187350973 | 0.03 |
ENST00000544130.1 |
ZC3H15 |
zinc finger CCCH-type containing 15 |
| chr7_+_114562172 | 0.03 |
ENST00000393486.1 ENST00000257724.3 |
MDFIC |
MyoD family inhibitor domain containing |
| chr1_-_162838551 | 0.03 |
ENST00000367910.1 ENST00000367912.2 ENST00000367911.2 |
C1orf110 |
chromosome 1 open reading frame 110 |
| chr22_-_19466732 | 0.03 |
ENST00000263202.10 ENST00000360834.4 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
| chr9_+_100174344 | 0.03 |
ENST00000422139.2 |
TDRD7 |
tudor domain containing 7 |
| chr12_-_57824739 | 0.03 |
ENST00000347140.3 ENST00000402412.1 |
R3HDM2 |
R3H domain containing 2 |
| chr1_+_225965518 | 0.03 |
ENST00000304786.7 ENST00000366839.4 ENST00000366838.1 |
SRP9 |
signal recognition particle 9kDa |
| chr14_-_102829051 | 0.03 |
ENST00000536961.2 ENST00000541568.2 ENST00000216756.6 |
CINP |
cyclin-dependent kinase 2 interacting protein |
| chr1_-_111217603 | 0.03 |
ENST00000369769.2 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr6_-_49755019 | 0.03 |
ENST00000304801.3 |
PGK2 |
phosphoglycerate kinase 2 |
| chr2_+_37423618 | 0.03 |
ENST00000402297.1 ENST00000397064.2 ENST00000406711.1 ENST00000392061.2 ENST00000397226.2 |
AC007390.5 |
CEBPZ antisense RNA 1 |
| chr22_-_30662828 | 0.03 |
ENST00000403463.1 ENST00000215781.2 |
OSM |
oncostatin M |
| chr8_+_145438870 | 0.03 |
ENST00000527931.1 |
FAM203B |
family with sequence similarity 203, member B |
| chr16_+_25123041 | 0.02 |
ENST00000399069.3 ENST00000380966.4 |
LCMT1 |
leucine carboxyl methyltransferase 1 |
| chrX_+_71354000 | 0.02 |
ENST00000510661.1 ENST00000535692.1 |
NHSL2 |
NHS-like 2 |
| chr12_-_56221330 | 0.02 |
ENST00000546837.1 |
RP11-762I7.5 |
Uncharacterized protein |
| chr19_-_51014460 | 0.02 |
ENST00000595669.1 |
JOSD2 |
Josephin domain containing 2 |
| chr7_+_94536898 | 0.02 |
ENST00000433360.1 ENST00000340694.4 ENST00000424654.1 |
PPP1R9A |
protein phosphatase 1, regulatory subunit 9A |
| chr12_+_54402790 | 0.02 |
ENST00000040584.4 |
HOXC8 |
homeobox C8 |
| chrX_+_71353499 | 0.02 |
ENST00000373677.1 |
NHSL2 |
NHS-like 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.2 | 0.7 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.2 | 0.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.4 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.1 | 0.3 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.1 | 0.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 1.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.5 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.4 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.3 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.2 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 1.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.7 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.7 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |