Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
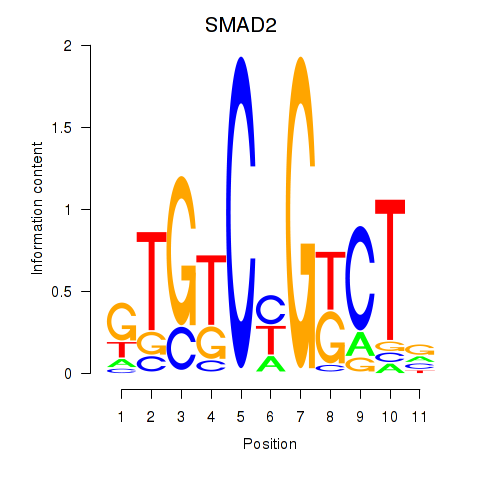
Results for SMAD2
Z-value: 0.64
Transcription factors associated with SMAD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD2
|
ENSG00000175387.11 | SMAD2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD2 | hg19_v2_chr18_-_45457192_45457278 | -0.49 | 2.2e-01 | Click! |
Activity profile of SMAD2 motif
Sorted Z-values of SMAD2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SMAD2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_159141397 | 1.02 |
ENST00000368124.4 ENST00000368125.4 ENST00000416746.1 |
CADM3 |
cell adhesion molecule 3 |
| chr12_+_53443963 | 0.59 |
ENST00000546602.1 ENST00000552570.1 ENST00000549700.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr21_+_47401650 | 0.48 |
ENST00000361866.3 |
COL6A1 |
collagen, type VI, alpha 1 |
| chr10_+_72972281 | 0.43 |
ENST00000335350.6 |
UNC5B |
unc-5 homolog B (C. elegans) |
| chr22_-_24641027 | 0.41 |
ENST00000398292.3 ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5 |
gamma-glutamyltransferase 5 |
| chr5_-_150521192 | 0.37 |
ENST00000523714.1 ENST00000521749.1 |
ANXA6 |
annexin A6 |
| chr17_-_15165854 | 0.35 |
ENST00000395936.1 ENST00000395938.2 |
PMP22 |
peripheral myelin protein 22 |
| chrX_-_107018969 | 0.34 |
ENST00000372383.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr6_-_152489484 | 0.31 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr12_+_53443680 | 0.30 |
ENST00000314250.6 ENST00000451358.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr16_-_3355645 | 0.30 |
ENST00000396862.1 ENST00000573608.1 |
TIGD7 |
tigger transposable element derived 7 |
| chr11_+_71238313 | 0.26 |
ENST00000398536.4 |
KRTAP5-7 |
keratin associated protein 5-7 |
| chr19_+_10527449 | 0.25 |
ENST00000592685.1 ENST00000380702.2 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr15_-_82338460 | 0.24 |
ENST00000558133.1 ENST00000329713.4 |
MEX3B |
mex-3 RNA binding family member B |
| chr11_-_5462744 | 0.23 |
ENST00000380211.1 |
OR51I1 |
olfactory receptor, family 51, subfamily I, member 1 |
| chr3_+_194406603 | 0.23 |
ENST00000329759.4 |
FAM43A |
family with sequence similarity 43, member A |
| chr12_-_29936731 | 0.23 |
ENST00000552618.1 ENST00000539277.1 ENST00000551659.1 |
TMTC1 |
transmembrane and tetratricopeptide repeat containing 1 |
| chr1_+_185703513 | 0.23 |
ENST00000271588.4 ENST00000367492.2 |
HMCN1 |
hemicentin 1 |
| chr8_-_95907423 | 0.22 |
ENST00000396133.3 ENST00000308108.4 |
CCNE2 |
cyclin E2 |
| chr20_+_34680620 | 0.19 |
ENST00000430276.1 ENST00000373950.2 ENST00000452261.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr7_-_100493744 | 0.18 |
ENST00000428317.1 ENST00000441605.1 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr16_+_31483374 | 0.18 |
ENST00000394863.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chr4_-_39640513 | 0.18 |
ENST00000511809.1 ENST00000505729.1 |
SMIM14 |
small integral membrane protein 14 |
| chr16_-_54320675 | 0.18 |
ENST00000329734.3 |
IRX3 |
iroquois homeobox 3 |
| chr19_-_51587502 | 0.17 |
ENST00000156499.2 ENST00000391802.1 |
KLK14 |
kallikrein-related peptidase 14 |
| chr16_-_17564738 | 0.17 |
ENST00000261381.6 |
XYLT1 |
xylosyltransferase I |
| chr4_-_39640700 | 0.17 |
ENST00000295958.5 |
SMIM14 |
small integral membrane protein 14 |
| chr7_-_100493482 | 0.16 |
ENST00000411582.1 ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr20_-_56284816 | 0.16 |
ENST00000395819.3 ENST00000341744.3 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chrX_-_107019181 | 0.16 |
ENST00000315660.4 ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr3_-_88108212 | 0.15 |
ENST00000482016.1 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr10_-_14372870 | 0.15 |
ENST00000357447.2 |
FRMD4A |
FERM domain containing 4A |
| chr5_+_140762268 | 0.14 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr6_+_41010293 | 0.13 |
ENST00000373161.1 ENST00000373158.2 ENST00000470917.1 |
TSPO2 |
translocator protein 2 |
| chr8_-_91657740 | 0.13 |
ENST00000422900.1 |
TMEM64 |
transmembrane protein 64 |
| chr21_+_46117087 | 0.13 |
ENST00000400365.3 |
KRTAP10-12 |
keratin associated protein 10-12 |
| chr3_+_197518100 | 0.13 |
ENST00000438796.2 ENST00000414675.2 ENST00000441090.2 ENST00000334859.4 ENST00000425562.2 |
LRCH3 |
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr8_+_95907993 | 0.13 |
ENST00000523378.1 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr15_+_75498739 | 0.12 |
ENST00000565074.1 |
C15orf39 |
chromosome 15 open reading frame 39 |
| chr6_+_6588902 | 0.12 |
ENST00000230568.4 |
LY86 |
lymphocyte antigen 86 |
| chr3_-_15374659 | 0.12 |
ENST00000426925.1 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
| chr4_-_113437328 | 0.12 |
ENST00000313341.3 |
NEUROG2 |
neurogenin 2 |
| chr6_+_132873832 | 0.11 |
ENST00000275200.1 |
TAAR8 |
trace amine associated receptor 8 |
| chr20_-_36889127 | 0.11 |
ENST00000279024.4 |
KIAA1755 |
KIAA1755 |
| chr6_+_39760129 | 0.11 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chrX_-_151143140 | 0.11 |
ENST00000393914.3 ENST00000370328.3 ENST00000370325.1 |
GABRE |
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chrX_-_48328631 | 0.11 |
ENST00000429543.1 ENST00000317669.5 |
SLC38A5 |
solute carrier family 38, member 5 |
| chr1_+_33722080 | 0.11 |
ENST00000483388.1 ENST00000539719.1 |
ZNF362 |
zinc finger protein 362 |
| chr6_+_39760783 | 0.10 |
ENST00000398904.2 ENST00000538976.1 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chrX_-_48328551 | 0.10 |
ENST00000376876.3 |
SLC38A5 |
solute carrier family 38, member 5 |
| chr6_+_147525541 | 0.10 |
ENST00000367481.3 ENST00000546097.1 |
STXBP5 |
syntaxin binding protein 5 (tomosyn) |
| chr1_-_211752073 | 0.10 |
ENST00000367001.4 |
SLC30A1 |
solute carrier family 30 (zinc transporter), member 1 |
| chr3_+_50316458 | 0.10 |
ENST00000316436.3 |
LSMEM2 |
leucine-rich single-pass membrane protein 2 |
| chr1_-_155271213 | 0.10 |
ENST00000342741.4 |
PKLR |
pyruvate kinase, liver and RBC |
| chr4_-_5890145 | 0.10 |
ENST00000397890.2 |
CRMP1 |
collapsin response mediator protein 1 |
| chr3_-_51937331 | 0.10 |
ENST00000310914.5 |
IQCF1 |
IQ motif containing F1 |
| chr7_-_105517021 | 0.10 |
ENST00000318724.4 ENST00000419735.3 |
ATXN7L1 |
ataxin 7-like 1 |
| chr17_+_68165657 | 0.10 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr9_+_140145713 | 0.10 |
ENST00000388931.3 ENST00000412566.1 |
C9orf173 |
chromosome 9 open reading frame 173 |
| chr11_-_33913708 | 0.09 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr22_-_31885514 | 0.09 |
ENST00000397525.1 |
EIF4ENIF1 |
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr16_+_53164833 | 0.09 |
ENST00000564845.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr7_-_105516923 | 0.09 |
ENST00000478915.1 |
ATXN7L1 |
ataxin 7-like 1 |
| chrX_-_154688276 | 0.09 |
ENST00000369445.2 |
F8A3 |
coagulation factor VIII-associated 3 |
| chr3_-_88108192 | 0.09 |
ENST00000309534.6 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr11_-_65150103 | 0.09 |
ENST00000294187.6 ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45 |
solute carrier family 25, member 45 |
| chr6_-_159466042 | 0.09 |
ENST00000338313.5 |
TAGAP |
T-cell activation RhoGTPase activating protein |
| chr11_-_75380165 | 0.09 |
ENST00000304771.3 |
MAP6 |
microtubule-associated protein 6 |
| chr3_+_88108381 | 0.09 |
ENST00000473136.1 |
RP11-159G9.5 |
Uncharacterized protein |
| chr20_+_43343517 | 0.08 |
ENST00000372865.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr12_-_89918522 | 0.08 |
ENST00000529983.2 |
GALNT4 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr11_-_75379612 | 0.08 |
ENST00000526740.1 |
MAP6 |
microtubule-associated protein 6 |
| chr1_+_36772691 | 0.08 |
ENST00000312808.4 ENST00000505871.1 |
SH3D21 |
SH3 domain containing 21 |
| chr9_+_125273081 | 0.08 |
ENST00000335302.5 |
OR1J2 |
olfactory receptor, family 1, subfamily J, member 2 |
| chr20_+_43343886 | 0.08 |
ENST00000190983.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr20_+_43343476 | 0.08 |
ENST00000372868.2 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr13_-_80915059 | 0.08 |
ENST00000377104.3 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
| chr1_-_1243252 | 0.08 |
ENST00000353662.3 |
ACAP3 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr1_-_201915590 | 0.07 |
ENST00000367288.4 |
LMOD1 |
leiomodin 1 (smooth muscle) |
| chr16_+_2198604 | 0.07 |
ENST00000210187.6 |
RAB26 |
RAB26, member RAS oncogene family |
| chr13_+_96743093 | 0.07 |
ENST00000376705.2 |
HS6ST3 |
heparan sulfate 6-O-sulfotransferase 3 |
| chr22_+_23213658 | 0.07 |
ENST00000390318.2 |
IGLV4-3 |
immunoglobulin lambda variable 4-3 |
| chr19_-_46288917 | 0.07 |
ENST00000537879.1 ENST00000596586.1 ENST00000595946.1 |
DMWD AC011530.4 |
dystrophia myotonica, WD repeat containing Uncharacterized protein |
| chr12_-_108991812 | 0.07 |
ENST00000392806.3 ENST00000547567.1 |
TMEM119 |
transmembrane protein 119 |
| chr1_-_59249732 | 0.07 |
ENST00000371222.2 |
JUN |
jun proto-oncogene |
| chr2_+_5832799 | 0.07 |
ENST00000322002.3 |
SOX11 |
SRY (sex determining region Y)-box 11 |
| chr5_+_14143728 | 0.07 |
ENST00000344204.4 ENST00000537187.1 |
TRIO |
trio Rho guanine nucleotide exchange factor |
| chr19_+_49977818 | 0.07 |
ENST00000594009.1 ENST00000595510.1 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
| chr3_-_71179699 | 0.07 |
ENST00000497355.1 |
FOXP1 |
forkhead box P1 |
| chr1_-_207119738 | 0.07 |
ENST00000356495.4 |
PIGR |
polymeric immunoglobulin receptor |
| chr19_+_15619299 | 0.07 |
ENST00000269703.3 |
CYP4F22 |
cytochrome P450, family 4, subfamily F, polypeptide 22 |
| chr9_+_33524240 | 0.06 |
ENST00000290943.6 |
ANKRD18B |
ankyrin repeat domain 18B |
| chr9_-_137809718 | 0.06 |
ENST00000371806.3 |
FCN1 |
ficolin (collagen/fibrinogen domain containing) 1 |
| chr22_-_38851205 | 0.06 |
ENST00000303592.3 |
KCNJ4 |
potassium inwardly-rectifying channel, subfamily J, member 4 |
| chr3_+_40547483 | 0.06 |
ENST00000420891.1 ENST00000314529.6 ENST00000418905.1 |
ZNF620 |
zinc finger protein 620 |
| chrX_+_153029633 | 0.06 |
ENST00000538966.1 ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3 |
plexin B3 |
| chr3_-_71179988 | 0.06 |
ENST00000491238.1 |
FOXP1 |
forkhead box P1 |
| chr11_+_2923423 | 0.06 |
ENST00000312221.5 |
SLC22A18 |
solute carrier family 22, member 18 |
| chr16_+_57702099 | 0.06 |
ENST00000333493.4 ENST00000327655.6 |
GPR97 |
G protein-coupled receptor 97 |
| chr9_-_73029540 | 0.06 |
ENST00000377126.2 |
KLF9 |
Kruppel-like factor 9 |
| chr20_+_44098385 | 0.06 |
ENST00000217425.5 ENST00000339946.3 |
WFDC2 |
WAP four-disulfide core domain 2 |
| chr11_-_65149422 | 0.06 |
ENST00000526432.1 ENST00000527174.1 |
SLC25A45 |
solute carrier family 25, member 45 |
| chr7_+_155250824 | 0.06 |
ENST00000297375.4 |
EN2 |
engrailed homeobox 2 |
| chr14_+_62162258 | 0.06 |
ENST00000337138.4 ENST00000394997.1 |
HIF1A |
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
| chr6_-_134861089 | 0.06 |
ENST00000606039.1 |
RP11-557H15.4 |
RP11-557H15.4 |
| chr8_-_142377367 | 0.06 |
ENST00000377741.3 |
GPR20 |
G protein-coupled receptor 20 |
| chr22_+_22764088 | 0.06 |
ENST00000390299.2 |
IGLV1-40 |
immunoglobulin lambda variable 1-40 |
| chr21_+_46066331 | 0.06 |
ENST00000334670.8 |
KRTAP10-11 |
keratin associated protein 10-11 |
| chr20_+_42143053 | 0.06 |
ENST00000373135.3 ENST00000444063.1 |
L3MBTL1 |
l(3)mbt-like 1 (Drosophila) |
| chr4_-_114682597 | 0.06 |
ENST00000394524.3 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr1_+_36771946 | 0.06 |
ENST00000373139.2 ENST00000453908.2 ENST00000426732.2 |
SH3D21 |
SH3 domain containing 21 |
| chr6_+_108487245 | 0.06 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chr11_+_74204395 | 0.06 |
ENST00000526036.1 |
AP001372.2 |
AP001372.2 |
| chr3_+_52529346 | 0.06 |
ENST00000321725.6 |
STAB1 |
stabilin 1 |
| chr19_-_44285401 | 0.06 |
ENST00000262888.3 |
KCNN4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr6_+_34482630 | 0.05 |
ENST00000538621.1 |
PACSIN1 |
protein kinase C and casein kinase substrate in neurons 1 |
| chr11_+_74951948 | 0.05 |
ENST00000562197.2 |
TPBGL |
trophoblast glycoprotein-like |
| chr10_-_75193308 | 0.05 |
ENST00000299432.2 |
MSS51 |
MSS51 mitochondrial translational activator |
| chr19_+_16999654 | 0.05 |
ENST00000248076.3 |
F2RL3 |
coagulation factor II (thrombin) receptor-like 3 |
| chr1_-_160040038 | 0.05 |
ENST00000368089.3 |
KCNJ10 |
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr17_-_1389419 | 0.05 |
ENST00000575158.1 |
MYO1C |
myosin IC |
| chr17_-_58469687 | 0.05 |
ENST00000590133.1 |
USP32 |
ubiquitin specific peptidase 32 |
| chr20_+_44098346 | 0.05 |
ENST00000372676.3 |
WFDC2 |
WAP four-disulfide core domain 2 |
| chr1_+_93913665 | 0.05 |
ENST00000271234.7 ENST00000370256.4 ENST00000260506.8 |
FNBP1L |
formin binding protein 1-like |
| chr8_-_130587254 | 0.05 |
ENST00000523151.1 |
CCDC26 |
coiled-coil domain containing 26 |
| chr20_+_30598231 | 0.05 |
ENST00000300415.8 ENST00000262659.8 |
CCM2L |
cerebral cavernous malformation 2-like |
| chr16_-_87417351 | 0.05 |
ENST00000311635.7 |
FBXO31 |
F-box protein 31 |
| chr5_+_70751442 | 0.05 |
ENST00000358731.4 ENST00000380675.2 |
BDP1 |
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr13_-_96705624 | 0.05 |
ENST00000376747.3 ENST00000376712.4 ENST00000397618.3 ENST00000376714.3 |
UGGT2 |
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr11_-_119211525 | 0.05 |
ENST00000528368.1 |
C1QTNF5 |
C1q and tumor necrosis factor related protein 5 |
| chr6_+_31730773 | 0.04 |
ENST00000415669.2 ENST00000425424.1 |
SAPCD1 |
suppressor APC domain containing 1 |
| chr4_+_7045042 | 0.04 |
ENST00000310074.7 ENST00000512388.1 |
TADA2B |
transcriptional adaptor 2B |
| chr12_+_110338323 | 0.04 |
ENST00000312777.5 ENST00000536408.2 |
TCHP |
trichoplein, keratin filament binding |
| chr17_-_10017864 | 0.04 |
ENST00000323816.4 |
GAS7 |
growth arrest-specific 7 |
| chr16_+_58059470 | 0.04 |
ENST00000219271.3 |
MMP15 |
matrix metallopeptidase 15 (membrane-inserted) |
| chr3_-_62359180 | 0.04 |
ENST00000283268.3 |
FEZF2 |
FEZ family zinc finger 2 |
| chrX_-_100183894 | 0.04 |
ENST00000328526.5 ENST00000372956.2 |
XKRX |
XK, Kell blood group complex subunit-related, X-linked |
| chr14_-_106494587 | 0.04 |
ENST00000390597.2 |
IGHV2-5 |
immunoglobulin heavy variable 2-5 |
| chr3_+_195447738 | 0.04 |
ENST00000447234.2 ENST00000320736.6 ENST00000436408.1 |
MUC20 |
mucin 20, cell surface associated |
| chr16_+_2286726 | 0.04 |
ENST00000382437.4 ENST00000569184.1 |
DNASE1L2 |
deoxyribonuclease I-like 2 |
| chr1_-_149908710 | 0.04 |
ENST00000439741.2 ENST00000361405.6 ENST00000406732.3 |
MTMR11 |
myotubularin related protein 11 |
| chr4_-_140477353 | 0.04 |
ENST00000406354.1 ENST00000506866.2 |
SETD7 |
SET domain containing (lysine methyltransferase) 7 |
| chr1_-_153508460 | 0.04 |
ENST00000462776.2 |
S100A6 |
S100 calcium binding protein A6 |
| chrX_-_99987088 | 0.04 |
ENST00000372981.1 ENST00000263033.5 |
SYTL4 |
synaptotagmin-like 4 |
| chr17_-_3571934 | 0.04 |
ENST00000225525.3 |
TAX1BP3 |
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr4_-_103748880 | 0.04 |
ENST00000453744.2 ENST00000349311.8 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr16_+_28889703 | 0.04 |
ENST00000357084.3 |
ATP2A1 |
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr1_+_15783222 | 0.04 |
ENST00000359621.4 |
CELA2A |
chymotrypsin-like elastase family, member 2A |
| chrX_-_47489244 | 0.04 |
ENST00000469388.1 ENST00000396992.3 ENST00000377005.2 |
CFP |
complement factor properdin |
| chr12_-_120554534 | 0.04 |
ENST00000538903.1 ENST00000534951.1 |
RAB35 |
RAB35, member RAS oncogene family |
| chr6_+_99282570 | 0.03 |
ENST00000328345.5 |
POU3F2 |
POU class 3 homeobox 2 |
| chr9_-_130829588 | 0.03 |
ENST00000373078.4 |
NAIF1 |
nuclear apoptosis inducing factor 1 |
| chr1_+_156163880 | 0.03 |
ENST00000359511.4 ENST00000423538.2 |
SLC25A44 |
solute carrier family 25, member 44 |
| chr9_+_5890802 | 0.03 |
ENST00000381477.3 ENST00000381476.1 ENST00000381471.1 |
MLANA |
melan-A |
| chr1_+_160097462 | 0.03 |
ENST00000447527.1 |
ATP1A2 |
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr21_-_36259445 | 0.03 |
ENST00000399240.1 |
RUNX1 |
runt-related transcription factor 1 |
| chr14_+_31343747 | 0.03 |
ENST00000216361.4 ENST00000396618.3 ENST00000475087.1 |
COCH |
cochlin |
| chr9_+_34990219 | 0.03 |
ENST00000541010.1 ENST00000454002.2 ENST00000545841.1 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr1_-_149908217 | 0.03 |
ENST00000369140.3 |
MTMR11 |
myotubularin related protein 11 |
| chr15_+_74610894 | 0.03 |
ENST00000558821.1 ENST00000268082.4 |
CCDC33 |
coiled-coil domain containing 33 |
| chr4_-_103749179 | 0.03 |
ENST00000502690.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr19_-_43383819 | 0.03 |
ENST00000312439.6 ENST00000403380.3 |
PSG1 |
pregnancy specific beta-1-glycoprotein 1 |
| chr13_+_114238997 | 0.03 |
ENST00000538138.1 ENST00000375370.5 |
TFDP1 |
transcription factor Dp-1 |
| chr2_-_220173685 | 0.03 |
ENST00000423636.2 ENST00000442029.1 ENST00000412847.1 |
PTPRN |
protein tyrosine phosphatase, receptor type, N |
| chr20_-_25062767 | 0.03 |
ENST00000429762.3 ENST00000444511.2 ENST00000376707.3 |
VSX1 |
visual system homeobox 1 |
| chr9_+_103790991 | 0.03 |
ENST00000374874.3 |
LPPR1 |
Lipid phosphate phosphatase-related protein type 1 |
| chr22_+_22735135 | 0.03 |
ENST00000390297.2 |
IGLV1-44 |
immunoglobulin lambda variable 1-44 |
| chr5_+_139493665 | 0.03 |
ENST00000331327.3 |
PURA |
purine-rich element binding protein A |
| chr12_-_10251539 | 0.03 |
ENST00000420265.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr4_-_164534657 | 0.03 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr6_-_159466136 | 0.03 |
ENST00000367066.3 ENST00000326965.6 |
TAGAP |
T-cell activation RhoGTPase activating protein |
| chr17_+_30593195 | 0.03 |
ENST00000431505.2 ENST00000269051.4 ENST00000538145.1 |
RHBDL3 |
rhomboid, veinlet-like 3 (Drosophila) |
| chr12_-_47473557 | 0.03 |
ENST00000321382.3 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr17_-_60142609 | 0.03 |
ENST00000397786.2 |
MED13 |
mediator complex subunit 13 |
| chr1_+_160085501 | 0.03 |
ENST00000361216.3 |
ATP1A2 |
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr20_+_36888551 | 0.03 |
ENST00000418004.1 ENST00000451435.1 |
BPI |
bactericidal/permeability-increasing protein |
| chr19_+_51645556 | 0.03 |
ENST00000601682.1 ENST00000317643.6 ENST00000305628.7 ENST00000600577.1 |
SIGLEC7 |
sialic acid binding Ig-like lectin 7 |
| chr21_-_38639813 | 0.02 |
ENST00000309117.6 ENST00000398998.1 |
DSCR3 |
Down syndrome critical region gene 3 |
| chrX_+_48660287 | 0.02 |
ENST00000444343.2 ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6 |
histone deacetylase 6 |
| chr2_+_168725458 | 0.02 |
ENST00000392690.3 |
B3GALT1 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr21_+_45993606 | 0.02 |
ENST00000400374.3 |
KRTAP10-4 |
keratin associated protein 10-4 |
| chr9_-_34523027 | 0.02 |
ENST00000399775.2 |
ENHO |
energy homeostasis associated |
| chr12_-_56583243 | 0.02 |
ENST00000550164.1 |
SMARCC2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr22_+_31518938 | 0.02 |
ENST00000412985.1 ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J |
inositol polyphosphate-5-phosphatase J |
| chr6_+_6588316 | 0.02 |
ENST00000379953.2 |
LY86 |
lymphocyte antigen 86 |
| chr19_-_42636543 | 0.02 |
ENST00000528894.4 ENST00000560804.2 ENST00000560558.1 ENST00000560398.1 ENST00000526816.2 |
POU2F2 |
POU class 2 homeobox 2 |
| chr12_-_47473425 | 0.02 |
ENST00000550413.1 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr20_-_33413416 | 0.02 |
ENST00000359003.2 |
NCOA6 |
nuclear receptor coactivator 6 |
| chr3_+_123813509 | 0.02 |
ENST00000460856.1 ENST00000240874.3 |
KALRN |
kalirin, RhoGEF kinase |
| chr16_+_28889801 | 0.02 |
ENST00000395503.4 |
ATP2A1 |
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr7_-_130080818 | 0.02 |
ENST00000343969.5 ENST00000541543.1 ENST00000489512.1 |
CEP41 |
centrosomal protein 41kDa |
| chr12_-_47473642 | 0.02 |
ENST00000266581.4 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr16_-_67997947 | 0.02 |
ENST00000537830.2 |
SLC12A4 |
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr5_+_64064748 | 0.02 |
ENST00000381070.3 ENST00000508024.1 |
CWC27 |
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr6_+_30689401 | 0.02 |
ENST00000396389.1 ENST00000396384.1 |
TUBB |
tubulin, beta class I |
| chr11_+_117857063 | 0.02 |
ENST00000227752.3 ENST00000541785.1 ENST00000545409.1 |
IL10RA |
interleukin 10 receptor, alpha |
| chr7_+_69064566 | 0.02 |
ENST00000403018.2 |
AUTS2 |
autism susceptibility candidate 2 |
| chr12_+_129178451 | 0.02 |
ENST00000537538.1 |
TMEM132C |
transmembrane protein 132C |
| chr12_+_68042495 | 0.02 |
ENST00000344096.3 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr4_+_93225550 | 0.02 |
ENST00000282020.4 |
GRID2 |
glutamate receptor, ionotropic, delta 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.2 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.2 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.4 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.1 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 1.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.0 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0097501 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.2 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.0 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.0 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.4 | GO:0043218 | compact myelin(GO:0043218) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.2 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.0 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |