Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
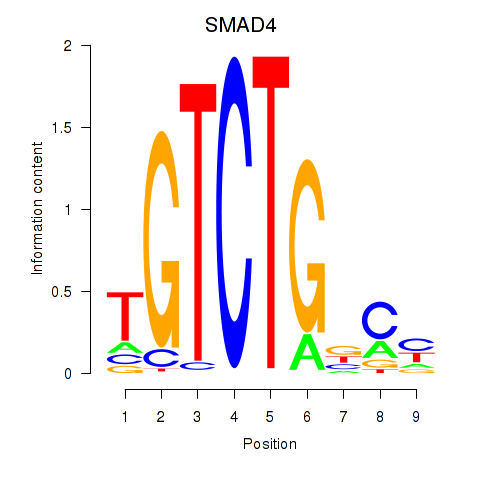
Results for SMAD4
Z-value: 0.58
Transcription factors associated with SMAD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD4
|
ENSG00000141646.9 | SMAD4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD4 | hg19_v2_chr18_+_48494361_48494426 | 0.55 | 1.6e-01 | Click! |
Activity profile of SMAD4 motif
Sorted Z-values of SMAD4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SMAD4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_24641027 | 1.06 |
ENST00000398292.3 ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5 |
gamma-glutamyltransferase 5 |
| chr1_+_159141397 | 1.01 |
ENST00000368124.4 ENST00000368125.4 ENST00000416746.1 |
CADM3 |
cell adhesion molecule 3 |
| chr5_+_156696362 | 0.71 |
ENST00000377576.3 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr13_+_102104980 | 0.67 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr7_+_94023873 | 0.66 |
ENST00000297268.6 |
COL1A2 |
collagen, type I, alpha 2 |
| chr14_-_92413353 | 0.61 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr13_+_102104952 | 0.61 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr1_+_159175201 | 0.60 |
ENST00000368121.2 |
DARC |
Duffy blood group, atypical chemokine receptor |
| chr5_-_111093340 | 0.58 |
ENST00000508870.1 |
NREP |
neuronal regeneration related protein |
| chr8_+_104384616 | 0.58 |
ENST00000520337.1 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr2_+_33359646 | 0.53 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359687 | 0.52 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr8_+_104383728 | 0.51 |
ENST00000330295.5 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr5_-_111092930 | 0.50 |
ENST00000257435.7 |
NREP |
neuronal regeneration related protein |
| chr11_-_70672645 | 0.49 |
ENST00000423696.2 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
| chr5_-_111093081 | 0.44 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr11_+_117070037 | 0.44 |
ENST00000392951.4 ENST00000525531.1 ENST00000278968.6 |
TAGLN |
transgelin |
| chr12_-_91572278 | 0.43 |
ENST00000425043.1 ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN |
decorin |
| chr8_-_120605194 | 0.41 |
ENST00000522167.1 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr5_-_111093406 | 0.40 |
ENST00000379671.3 |
NREP |
neuronal regeneration related protein |
| chr9_+_120466610 | 0.40 |
ENST00000394487.4 |
TLR4 |
toll-like receptor 4 |
| chr5_-_168727786 | 0.38 |
ENST00000332966.8 |
SLIT3 |
slit homolog 3 (Drosophila) |
| chr5_-_124081008 | 0.37 |
ENST00000306315.5 |
ZNF608 |
zinc finger protein 608 |
| chr11_-_118550346 | 0.36 |
ENST00000530256.1 |
TREH |
trehalase (brush-border membrane glycoprotein) |
| chr22_+_31003133 | 0.36 |
ENST00000405742.3 |
TCN2 |
transcobalamin II |
| chr12_+_53443963 | 0.34 |
ENST00000546602.1 ENST00000552570.1 ENST00000549700.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr10_+_31610064 | 0.34 |
ENST00000446923.2 ENST00000559476.1 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr17_-_67138015 | 0.33 |
ENST00000284425.2 ENST00000590645.1 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chrX_+_100743031 | 0.33 |
ENST00000423738.3 |
ARMCX4 |
armadillo repeat containing, X-linked 4 |
| chr5_-_121413974 | 0.32 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chr5_-_168727713 | 0.32 |
ENST00000404867.3 |
SLIT3 |
slit homolog 3 (Drosophila) |
| chr12_+_1738363 | 0.32 |
ENST00000397196.2 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
| chr22_+_31003190 | 0.31 |
ENST00000407817.3 |
TCN2 |
transcobalamin II |
| chr5_-_150521192 | 0.31 |
ENST00000523714.1 ENST00000521749.1 |
ANXA6 |
annexin A6 |
| chr11_-_118550375 | 0.31 |
ENST00000525958.1 ENST00000264029.4 ENST00000397925.1 ENST00000529101.1 |
TREH |
trehalase (brush-border membrane glycoprotein) |
| chr22_+_31002779 | 0.30 |
ENST00000215838.3 |
TCN2 |
transcobalamin II |
| chr3_+_154797877 | 0.30 |
ENST00000462745.1 ENST00000493237.1 |
MME |
membrane metallo-endopeptidase |
| chr11_-_111794446 | 0.30 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr5_-_149535421 | 0.29 |
ENST00000261799.4 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
| chr1_-_32827682 | 0.29 |
ENST00000432622.1 |
FAM229A |
family with sequence similarity 229, member A |
| chr19_+_37342547 | 0.29 |
ENST00000331800.4 ENST00000586646.1 |
ZNF345 |
zinc finger protein 345 |
| chr5_-_168728103 | 0.28 |
ENST00000519560.1 |
SLIT3 |
slit homolog 3 (Drosophila) |
| chr14_-_106406090 | 0.28 |
ENST00000390593.2 |
IGHV6-1 |
immunoglobulin heavy variable 6-1 |
| chrX_-_154250989 | 0.28 |
ENST00000360256.4 |
F8 |
coagulation factor VIII, procoagulant component |
| chr16_+_2880369 | 0.28 |
ENST00000572863.1 |
ZG16B |
zymogen granule protein 16B |
| chr13_+_76378305 | 0.28 |
ENST00000526371.1 ENST00000526528.1 |
LMO7 |
LIM domain 7 |
| chr2_+_30454390 | 0.27 |
ENST00000395323.3 ENST00000406087.1 ENST00000404397.1 |
LBH |
limb bud and heart development |
| chr15_-_82338460 | 0.26 |
ENST00000558133.1 ENST00000329713.4 |
MEX3B |
mex-3 RNA binding family member B |
| chr10_+_94451574 | 0.26 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chr17_-_19290117 | 0.25 |
ENST00000497081.2 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr4_-_100242549 | 0.25 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr11_+_86511549 | 0.25 |
ENST00000533902.2 |
PRSS23 |
protease, serine, 23 |
| chr19_+_49617581 | 0.25 |
ENST00000391864.3 |
LIN7B |
lin-7 homolog B (C. elegans) |
| chr2_-_45236540 | 0.25 |
ENST00000303077.6 |
SIX2 |
SIX homeobox 2 |
| chr1_-_153522562 | 0.25 |
ENST00000368714.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr3_+_126243126 | 0.25 |
ENST00000319340.2 |
CHST13 |
carbohydrate (chondroitin 4) sulfotransferase 13 |
| chr16_+_2880157 | 0.25 |
ENST00000382280.3 |
ZG16B |
zymogen granule protein 16B |
| chr13_+_76210448 | 0.24 |
ENST00000377499.5 |
LMO7 |
LIM domain 7 |
| chr19_+_36359341 | 0.24 |
ENST00000221891.4 |
APLP1 |
amyloid beta (A4) precursor-like protein 1 |
| chr1_-_31230650 | 0.23 |
ENST00000294507.3 |
LAPTM5 |
lysosomal protein transmembrane 5 |
| chr5_+_148521136 | 0.23 |
ENST00000506113.1 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr5_+_148521046 | 0.23 |
ENST00000326685.7 ENST00000356541.3 ENST00000309868.7 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr22_+_44319648 | 0.23 |
ENST00000423180.2 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
| chr2_+_217082311 | 0.23 |
ENST00000597904.1 |
RP11-566E18.3 |
RP11-566E18.3 |
| chr5_+_148521381 | 0.23 |
ENST00000504238.1 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr7_-_100493482 | 0.22 |
ENST00000411582.1 ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr12_+_53443680 | 0.22 |
ENST00000314250.6 ENST00000451358.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr3_+_46616017 | 0.22 |
ENST00000542931.1 |
TDGF1 |
teratocarcinoma-derived growth factor 1 |
| chr19_+_10527449 | 0.22 |
ENST00000592685.1 ENST00000380702.2 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr5_+_82767583 | 0.22 |
ENST00000512590.2 ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN |
versican |
| chr17_-_19290483 | 0.22 |
ENST00000395592.2 ENST00000299610.4 |
MFAP4 |
microfibrillar-associated protein 4 |
| chr6_-_56707943 | 0.22 |
ENST00000370769.4 ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST |
dystonin |
| chr16_+_2880296 | 0.22 |
ENST00000571723.1 |
ZG16B |
zymogen granule protein 16B |
| chr1_-_153599732 | 0.21 |
ENST00000392623.1 |
S100A13 |
S100 calcium binding protein A13 |
| chr2_+_217524323 | 0.21 |
ENST00000456764.1 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr15_+_67420441 | 0.21 |
ENST00000558894.1 |
SMAD3 |
SMAD family member 3 |
| chrX_-_151143140 | 0.21 |
ENST00000393914.3 ENST00000370328.3 ENST00000370325.1 |
GABRE |
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr22_+_44319619 | 0.21 |
ENST00000216180.3 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
| chr4_+_671711 | 0.21 |
ENST00000400159.2 |
MYL5 |
myosin, light chain 5, regulatory |
| chr3_+_106959530 | 0.20 |
ENST00000466734.1 ENST00000463143.1 ENST00000490441.1 |
LINC00883 |
long intergenic non-protein coding RNA 883 |
| chr13_+_76378357 | 0.20 |
ENST00000489941.2 ENST00000525373.1 |
LMO7 |
LIM domain 7 |
| chr1_-_232697304 | 0.20 |
ENST00000366630.1 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr4_+_126237554 | 0.20 |
ENST00000394329.3 |
FAT4 |
FAT atypical cadherin 4 |
| chr11_-_111781554 | 0.20 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr5_+_126626498 | 0.20 |
ENST00000503335.2 ENST00000508365.1 ENST00000418761.2 ENST00000274473.6 |
MEGF10 |
multiple EGF-like-domains 10 |
| chr9_-_21305312 | 0.20 |
ENST00000259555.4 |
IFNA5 |
interferon, alpha 5 |
| chr12_+_75874460 | 0.20 |
ENST00000266659.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr3_-_48594248 | 0.19 |
ENST00000545984.1 ENST00000232375.3 ENST00000416568.1 ENST00000383734.2 ENST00000541519.1 ENST00000412035.1 |
PFKFB4 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr11_-_111781454 | 0.19 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr19_+_51628165 | 0.19 |
ENST00000250360.3 ENST00000440804.3 |
SIGLEC9 |
sialic acid binding Ig-like lectin 9 |
| chr1_+_210406121 | 0.19 |
ENST00000367012.3 |
SERTAD4 |
SERTA domain containing 4 |
| chr7_-_100493744 | 0.19 |
ENST00000428317.1 ENST00000441605.1 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr20_+_34680620 | 0.19 |
ENST00000430276.1 ENST00000373950.2 ENST00000452261.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr12_+_75874984 | 0.19 |
ENST00000550491.1 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr10_+_102891048 | 0.19 |
ENST00000467928.2 |
TLX1 |
T-cell leukemia homeobox 1 |
| chr11_-_111781610 | 0.18 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr12_-_31744031 | 0.18 |
ENST00000389082.5 |
DENND5B |
DENN/MADD domain containing 5B |
| chr3_-_48632593 | 0.18 |
ENST00000454817.1 ENST00000328333.8 |
COL7A1 |
collagen, type VII, alpha 1 |
| chr5_-_127674883 | 0.18 |
ENST00000507835.1 |
FBN2 |
fibrillin 2 |
| chr3_-_114477962 | 0.18 |
ENST00000471418.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr10_+_91087651 | 0.18 |
ENST00000371818.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr16_-_31076332 | 0.18 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chr9_+_99690592 | 0.18 |
ENST00000354649.3 |
NUTM2G |
NUT family member 2G |
| chr3_+_141103634 | 0.18 |
ENST00000507722.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr10_+_81892347 | 0.18 |
ENST00000372267.2 |
PLAC9 |
placenta-specific 9 |
| chr14_+_21538429 | 0.18 |
ENST00000298694.4 ENST00000555038.1 |
ARHGEF40 |
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr13_+_31309645 | 0.18 |
ENST00000380490.3 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
| chr9_-_139891165 | 0.17 |
ENST00000494426.1 |
CLIC3 |
chloride intracellular channel 3 |
| chr12_+_75874580 | 0.17 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr14_+_21538517 | 0.17 |
ENST00000298693.3 |
ARHGEF40 |
Rho guanine nucleotide exchange factor (GEF) 40 |
| chrX_+_54835493 | 0.17 |
ENST00000396224.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr14_+_21156915 | 0.17 |
ENST00000397990.4 ENST00000555597.1 |
ANG RNASE4 |
angiogenin, ribonuclease, RNase A family, 5 ribonuclease, RNase A family, 4 |
| chr19_+_41768561 | 0.17 |
ENST00000599719.1 ENST00000601309.1 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr1_+_110210644 | 0.17 |
ENST00000369831.2 ENST00000442650.1 ENST00000369827.3 ENST00000460717.3 ENST00000241337.4 ENST00000467579.3 ENST00000414179.2 ENST00000369829.2 |
GSTM2 |
glutathione S-transferase mu 2 (muscle) |
| chr11_-_27722021 | 0.17 |
ENST00000356660.4 ENST00000418212.1 ENST00000533246.1 |
BDNF |
brain-derived neurotrophic factor |
| chr1_+_110036728 | 0.17 |
ENST00000369868.3 ENST00000430195.2 |
CYB561D1 |
cytochrome b561 family, member D1 |
| chr10_-_62704005 | 0.17 |
ENST00000337910.5 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chr4_-_95264008 | 0.16 |
ENST00000295256.5 |
HPGDS |
hematopoietic prostaglandin D synthase |
| chr5_+_140529630 | 0.16 |
ENST00000543635.1 |
PCDHB6 |
protocadherin beta 6 |
| chr22_+_31090793 | 0.16 |
ENST00000332585.6 ENST00000382310.3 ENST00000446658.2 |
OSBP2 |
oxysterol binding protein 2 |
| chr6_+_41010293 | 0.16 |
ENST00000373161.1 ENST00000373158.2 ENST00000470917.1 |
TSPO2 |
translocator protein 2 |
| chr15_-_83316254 | 0.16 |
ENST00000567678.1 ENST00000450751.2 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr5_+_149569520 | 0.16 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chrX_+_153029633 | 0.16 |
ENST00000538966.1 ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3 |
plexin B3 |
| chr11_-_65150103 | 0.16 |
ENST00000294187.6 ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45 |
solute carrier family 25, member 45 |
| chr5_-_124080203 | 0.16 |
ENST00000504926.1 |
ZNF608 |
zinc finger protein 608 |
| chr11_+_67777751 | 0.16 |
ENST00000316367.6 ENST00000007633.8 ENST00000342456.6 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
| chr7_-_141646726 | 0.16 |
ENST00000438351.1 ENST00000439991.1 ENST00000551012.2 ENST00000546910.1 |
CLEC5A |
C-type lectin domain family 5, member A |
| chr8_-_57472154 | 0.16 |
ENST00000499425.1 ENST00000523664.1 ENST00000518943.1 ENST00000524338.1 |
LINC00968 |
long intergenic non-protein coding RNA 968 |
| chr11_-_3186494 | 0.16 |
ENST00000389989.3 ENST00000542243.1 |
OSBPL5 |
oxysterol binding protein-like 5 |
| chr15_-_43910998 | 0.15 |
ENST00000450892.2 |
STRC |
stereocilin |
| chr7_+_20687017 | 0.15 |
ENST00000258738.6 |
ABCB5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr1_+_110036699 | 0.15 |
ENST00000496961.1 ENST00000533024.1 ENST00000310611.4 ENST00000527072.1 ENST00000420578.2 ENST00000528785.1 |
CYB561D1 |
cytochrome b561 family, member D1 |
| chr14_+_103566481 | 0.15 |
ENST00000380069.3 |
EXOC3L4 |
exocyst complex component 3-like 4 |
| chr19_-_46285736 | 0.15 |
ENST00000291270.4 ENST00000447742.2 ENST00000354227.5 |
DMPK |
dystrophia myotonica-protein kinase |
| chr22_+_23077065 | 0.15 |
ENST00000390310.2 |
IGLV2-18 |
immunoglobulin lambda variable 2-18 |
| chr19_-_46285646 | 0.15 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr19_-_47922373 | 0.15 |
ENST00000559524.1 ENST00000557833.1 ENST00000558555.1 ENST00000561293.1 ENST00000441740.2 |
MEIS3 |
Meis homeobox 3 |
| chr17_-_76870126 | 0.15 |
ENST00000586057.1 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr11_+_64879317 | 0.15 |
ENST00000526809.1 ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2 |
transmembrane 7 superfamily member 2 |
| chr1_-_46598371 | 0.15 |
ENST00000372006.1 ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr2_-_127963343 | 0.15 |
ENST00000335247.7 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr20_-_56265680 | 0.15 |
ENST00000414037.1 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr11_+_86511569 | 0.15 |
ENST00000441050.1 |
PRSS23 |
protease, serine, 23 |
| chr22_+_31518938 | 0.14 |
ENST00000412985.1 ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J |
inositol polyphosphate-5-phosphatase J |
| chr11_+_43964055 | 0.14 |
ENST00000528572.1 |
C11orf96 |
chromosome 11 open reading frame 96 |
| chr2_-_175711133 | 0.14 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chrX_-_51812268 | 0.14 |
ENST00000486010.1 ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B |
melanoma antigen family D, 4B |
| chr1_-_154461642 | 0.14 |
ENST00000555188.1 |
SHE |
Src homology 2 domain containing E |
| chr6_+_116892530 | 0.14 |
ENST00000466444.2 ENST00000368590.5 ENST00000392526.1 |
RWDD1 |
RWD domain containing 1 |
| chr6_+_39760129 | 0.14 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr7_-_150675372 | 0.14 |
ENST00000262186.5 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr3_+_9958870 | 0.14 |
ENST00000413608.1 |
IL17RC |
interleukin 17 receptor C |
| chr19_+_49617609 | 0.14 |
ENST00000221459.2 ENST00000486217.2 |
LIN7B |
lin-7 homolog B (C. elegans) |
| chr16_-_31076273 | 0.14 |
ENST00000426488.2 |
ZNF668 |
zinc finger protein 668 |
| chr8_+_38586068 | 0.14 |
ENST00000443286.2 ENST00000520340.1 ENST00000518415.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr10_+_72972281 | 0.14 |
ENST00000335350.6 |
UNC5B |
unc-5 homolog B (C. elegans) |
| chrX_-_48328631 | 0.14 |
ENST00000429543.1 ENST00000317669.5 |
SLC38A5 |
solute carrier family 38, member 5 |
| chr3_+_9958758 | 0.14 |
ENST00000383812.4 ENST00000438091.1 ENST00000295981.3 ENST00000436503.1 ENST00000403601.3 ENST00000416074.2 ENST00000455057.1 |
IL17RC |
interleukin 17 receptor C |
| chr2_+_201170703 | 0.14 |
ENST00000358677.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr22_-_38851205 | 0.14 |
ENST00000303592.3 |
KCNJ4 |
potassium inwardly-rectifying channel, subfamily J, member 4 |
| chrX_-_48328551 | 0.14 |
ENST00000376876.3 |
SLC38A5 |
solute carrier family 38, member 5 |
| chr15_+_43803143 | 0.14 |
ENST00000382031.1 |
MAP1A |
microtubule-associated protein 1A |
| chr3_+_50316458 | 0.14 |
ENST00000316436.3 |
LSMEM2 |
leucine-rich single-pass membrane protein 2 |
| chr8_+_38585704 | 0.13 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr16_+_31483374 | 0.13 |
ENST00000394863.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chr15_-_52944231 | 0.13 |
ENST00000546305.2 |
FAM214A |
family with sequence similarity 214, member A |
| chr11_-_5462744 | 0.13 |
ENST00000380211.1 |
OR51I1 |
olfactory receptor, family 51, subfamily I, member 1 |
| chr9_+_72658490 | 0.13 |
ENST00000377182.4 |
MAMDC2 |
MAM domain containing 2 |
| chr10_-_28287968 | 0.13 |
ENST00000305242.5 |
ARMC4 |
armadillo repeat containing 4 |
| chrX_-_48931648 | 0.13 |
ENST00000376386.3 ENST00000376390.4 |
PRAF2 |
PRA1 domain family, member 2 |
| chr1_-_46598284 | 0.13 |
ENST00000423209.1 ENST00000262741.5 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr10_+_26505594 | 0.13 |
ENST00000259271.3 |
GAD2 |
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) |
| chr22_+_22930626 | 0.13 |
ENST00000390302.2 |
IGLV2-33 |
immunoglobulin lambda variable 2-33 (non-functional) |
| chr4_+_74718906 | 0.13 |
ENST00000226524.3 |
PF4V1 |
platelet factor 4 variant 1 |
| chr16_-_54320675 | 0.13 |
ENST00000329734.3 |
IRX3 |
iroquois homeobox 3 |
| chr3_+_187086120 | 0.13 |
ENST00000259030.2 |
RTP4 |
receptor (chemosensory) transporter protein 4 |
| chr9_+_140119618 | 0.13 |
ENST00000359069.2 |
C9orf169 |
chromosome 9 open reading frame 169 |
| chr2_+_201997676 | 0.13 |
ENST00000462763.1 ENST00000479953.2 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr2_+_42721689 | 0.13 |
ENST00000405592.1 |
MTA3 |
metastasis associated 1 family, member 3 |
| chr4_-_39640700 | 0.13 |
ENST00000295958.5 |
SMIM14 |
small integral membrane protein 14 |
| chrX_-_102757802 | 0.13 |
ENST00000372633.1 |
RAB40A |
RAB40A, member RAS oncogene family |
| chr19_+_48898132 | 0.12 |
ENST00000263269.3 |
GRIN2D |
glutamate receptor, ionotropic, N-methyl D-aspartate 2D |
| chr1_+_180165672 | 0.12 |
ENST00000443059.1 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
| chr4_+_55524085 | 0.12 |
ENST00000412167.2 ENST00000288135.5 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr1_-_238649319 | 0.12 |
ENST00000400946.2 |
RP11-371I1.2 |
long intergenic non-protein coding RNA 1139 |
| chr20_+_43538756 | 0.12 |
ENST00000537323.1 ENST00000217073.2 |
PABPC1L |
poly(A) binding protein, cytoplasmic 1-like |
| chr6_+_84222194 | 0.12 |
ENST00000536636.1 |
PRSS35 |
protease, serine, 35 |
| chr9_+_34652164 | 0.12 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chr6_+_108487245 | 0.12 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chr16_+_2880254 | 0.12 |
ENST00000570670.1 |
ZG16B |
zymogen granule protein 16B |
| chr1_+_11751748 | 0.12 |
ENST00000294485.5 |
DRAXIN |
dorsal inhibitory axon guidance protein |
| chr21_+_39628655 | 0.12 |
ENST00000398925.1 ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr20_+_43538692 | 0.12 |
ENST00000217074.4 ENST00000255136.3 |
PABPC1L |
poly(A) binding protein, cytoplasmic 1-like |
| chrX_+_54947229 | 0.12 |
ENST00000442098.1 ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO |
trophinin |
| chr19_+_41117770 | 0.12 |
ENST00000601032.1 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr10_+_81891416 | 0.12 |
ENST00000372270.2 |
PLAC9 |
placenta-specific 9 |
| chr22_-_31503490 | 0.12 |
ENST00000400299.2 |
SELM |
Selenoprotein M |
| chr11_-_118213360 | 0.12 |
ENST00000529594.1 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
| chr6_-_87804815 | 0.12 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.2 | 1.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.2 | 1.0 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 0.4 | GO:0070428 | negative regulation of interleukin-23 production(GO:0032707) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 | 1.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.5 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 1.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.3 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.1 | 0.3 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.2 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.3 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular response to UV-A(GO:0071492) |
| 0.1 | 0.2 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.1 | 0.7 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.3 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.1 | 0.2 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.3 | GO:0070458 | cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.7 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.0 | 0.8 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.0 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 0.1 | GO:0038162 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.4 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.0 | 0.1 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.2 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.2 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.3 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.3 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.6 | GO:0030208 | dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:1901295 | positive regulation of ephrin receptor signaling pathway(GO:1901189) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:1902228 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.0 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.0 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 1.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.0 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 | 0.1 | GO:2000737 | negative regulation of stem cell differentiation(GO:2000737) |
| 0.0 | 0.3 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.0 | GO:1903524 | regulation of vasoconstriction(GO:0019229) positive regulation of vasoconstriction(GO:0045907) positive regulation of blood circulation(GO:1903524) |
| 0.0 | 0.1 | GO:1902023 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:1904404 | transcription factor catabolic process(GO:0036369) cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0060621 | negative regulation of cholesterol import(GO:0060621) negative regulation of sterol import(GO:2000910) |
| 0.0 | 0.3 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.0 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.0 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.0 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.0 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.0 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:2000452 | CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:0035698) positive regulation of necroptotic process(GO:0060545) regulation of CD8-positive, alpha-beta cytotoxic T cell extravasation(GO:2000452) |
| 0.0 | 0.4 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.1 | GO:0002251 | organ or tissue specific immune response(GO:0002251) mucosal immune response(GO:0002385) |
| 0.0 | 0.4 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.0 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.2 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.0 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.9 | GO:0001895 | retina homeostasis(GO:0001895) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.9 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.0 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.0 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.5 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 1.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 1.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.2 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.1 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.0 | GO:0043125 | ErbB-2 class receptor binding(GO:0005176) ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.0 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.0 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.0 | GO:0033300 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) dehydroascorbic acid transporter activity(GO:0033300) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 3.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |