Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
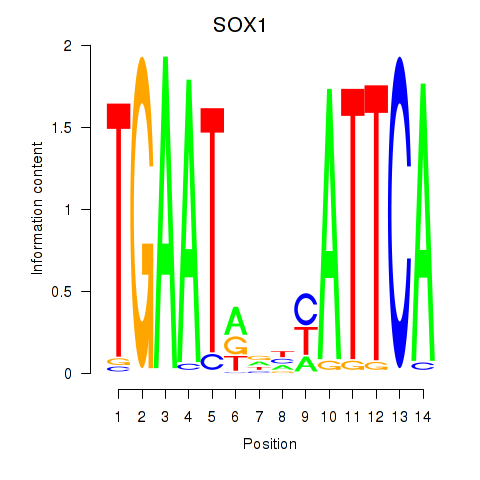
Results for SOX1
Z-value: 0.32
Transcription factors associated with SOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX1
|
ENSG00000182968.3 | SOX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX1 | hg19_v2_chr13_+_112721913_112721913 | -0.05 | 9.1e-01 | Click! |
Activity profile of SOX1 motif
Sorted Z-values of SOX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_189840223 | 0.68 |
ENST00000427335.2 |
LEPREL1 |
leprecan-like 1 |
| chr3_+_136676851 | 0.45 |
ENST00000309741.5 |
IL20RB |
interleukin 20 receptor beta |
| chr3_+_136676707 | 0.44 |
ENST00000329582.4 |
IL20RB |
interleukin 20 receptor beta |
| chr16_+_12058961 | 0.40 |
ENST00000053243.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr16_+_12059050 | 0.38 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr1_+_152957707 | 0.32 |
ENST00000368762.1 |
SPRR1A |
small proline-rich protein 1A |
| chr4_-_74486217 | 0.29 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr2_+_102615416 | 0.27 |
ENST00000393414.2 |
IL1R2 |
interleukin 1 receptor, type II |
| chr17_-_39646116 | 0.24 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr1_+_70876926 | 0.23 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr8_-_110620284 | 0.23 |
ENST00000529690.1 |
SYBU |
syntabulin (syntaxin-interacting) |
| chr9_-_104249400 | 0.22 |
ENST00000374848.3 |
TMEM246 |
transmembrane protein 246 |
| chr13_+_109248500 | 0.19 |
ENST00000356711.2 |
MYO16 |
myosin XVI |
| chr17_-_29624343 | 0.17 |
ENST00000247271.4 |
OMG |
oligodendrocyte myelin glycoprotein |
| chr20_-_14318248 | 0.16 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr11_+_55029628 | 0.14 |
ENST00000417545.2 |
TRIM48 |
tripartite motif containing 48 |
| chr11_-_57177586 | 0.13 |
ENST00000529411.1 |
RP11-872D17.8 |
Uncharacterized protein |
| chr8_+_145133493 | 0.13 |
ENST00000316052.5 ENST00000525936.1 |
EXOSC4 |
exosome component 4 |
| chr12_+_115800817 | 0.13 |
ENST00000547948.1 |
RP11-116D17.1 |
HCG2038717; Uncharacterized protein |
| chr1_-_200379129 | 0.12 |
ENST00000367353.1 |
ZNF281 |
zinc finger protein 281 |
| chr4_-_74486109 | 0.12 |
ENST00000395777.2 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr9_-_95166884 | 0.12 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr4_-_74486347 | 0.12 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr9_-_95166841 | 0.11 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr1_-_200379180 | 0.11 |
ENST00000294740.3 |
ZNF281 |
zinc finger protein 281 |
| chr6_+_26183958 | 0.11 |
ENST00000356530.3 |
HIST1H2BE |
histone cluster 1, H2be |
| chr1_+_32674675 | 0.10 |
ENST00000409358.1 |
DCDC2B |
doublecortin domain containing 2B |
| chr1_+_46805832 | 0.10 |
ENST00000474844.1 |
NSUN4 |
NOP2/Sun domain family, member 4 |
| chr1_+_152975488 | 0.10 |
ENST00000542696.1 |
SPRR3 |
small proline-rich protein 3 |
| chr5_-_16916624 | 0.10 |
ENST00000513882.1 |
MYO10 |
myosin X |
| chr12_-_120763739 | 0.09 |
ENST00000549767.1 |
PLA2G1B |
phospholipase A2, group IB (pancreas) |
| chr2_-_220083076 | 0.09 |
ENST00000295750.4 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr11_-_133715394 | 0.09 |
ENST00000299140.3 ENST00000532889.1 |
SPATA19 |
spermatogenesis associated 19 |
| chr12_-_10151773 | 0.09 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr3_-_33686925 | 0.09 |
ENST00000485378.2 ENST00000313350.6 ENST00000487200.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr8_+_110551925 | 0.09 |
ENST00000395785.2 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
| chr2_-_163175133 | 0.08 |
ENST00000421365.2 ENST00000263642.2 |
IFIH1 |
interferon induced with helicase C domain 1 |
| chr1_+_19578033 | 0.08 |
ENST00000330263.4 |
MRTO4 |
mRNA turnover 4 homolog (S. cerevisiae) |
| chr4_+_70861647 | 0.08 |
ENST00000246895.4 ENST00000381060.2 |
STATH |
statherin |
| chr4_-_170924888 | 0.08 |
ENST00000502832.1 ENST00000393704.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr1_+_240255166 | 0.07 |
ENST00000319653.9 |
FMN2 |
formin 2 |
| chr14_+_22356029 | 0.07 |
ENST00000390437.2 |
TRAV12-2 |
T cell receptor alpha variable 12-2 |
| chrX_+_129040122 | 0.07 |
ENST00000394422.3 ENST00000371051.5 |
UTP14A |
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr2_-_209054709 | 0.07 |
ENST00000449053.1 ENST00000451346.1 ENST00000341287.4 |
C2orf80 |
chromosome 2 open reading frame 80 |
| chr3_+_158362299 | 0.07 |
ENST00000478576.1 ENST00000264263.5 ENST00000464732.1 |
GFM1 |
G elongation factor, mitochondrial 1 |
| chr6_-_71666732 | 0.07 |
ENST00000230053.6 |
B3GAT2 |
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr7_-_142176790 | 0.07 |
ENST00000390369.2 |
TRBV7-4 |
T cell receptor beta variable 7-4 (gene/pseudogene) |
| chr9_-_2844058 | 0.07 |
ENST00000397885.2 |
KIAA0020 |
KIAA0020 |
| chr17_-_38821373 | 0.06 |
ENST00000394052.3 |
KRT222 |
keratin 222 |
| chr20_+_31805131 | 0.06 |
ENST00000375454.3 ENST00000375452.3 |
BPIFA3 |
BPI fold containing family A, member 3 |
| chrX_+_129040094 | 0.06 |
ENST00000425117.2 |
UTP14A |
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
| chr11_-_123756334 | 0.06 |
ENST00000528595.1 ENST00000375026.2 |
TMEM225 |
transmembrane protein 225 |
| chr1_-_200379104 | 0.06 |
ENST00000367352.3 |
ZNF281 |
zinc finger protein 281 |
| chr3_-_58200398 | 0.06 |
ENST00000318316.3 ENST00000460422.1 ENST00000483681.1 |
DNASE1L3 |
deoxyribonuclease I-like 3 |
| chr10_+_78078088 | 0.05 |
ENST00000496424.2 |
C10orf11 |
chromosome 10 open reading frame 11 |
| chr11_-_47400062 | 0.05 |
ENST00000533030.1 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr1_+_174769006 | 0.05 |
ENST00000489615.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr1_-_169455169 | 0.05 |
ENST00000367804.4 ENST00000236137.5 |
SLC19A2 |
solute carrier family 19 (thiamine transporter), member 2 |
| chr11_-_5255861 | 0.05 |
ENST00000380299.3 |
HBD |
hemoglobin, delta |
| chr1_-_19578003 | 0.05 |
ENST00000375199.3 ENST00000375208.3 ENST00000356068.2 ENST00000477853.1 |
EMC1 |
ER membrane protein complex subunit 1 |
| chr8_-_86290333 | 0.05 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr11_-_47399942 | 0.05 |
ENST00000227163.4 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr7_-_36634181 | 0.05 |
ENST00000538464.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
| chr19_+_35842445 | 0.05 |
ENST00000246553.2 |
FFAR1 |
free fatty acid receptor 1 |
| chr11_-_113577052 | 0.05 |
ENST00000540540.1 ENST00000545579.1 ENST00000538955.1 ENST00000299882.5 |
TMPRSS5 |
transmembrane protease, serine 5 |
| chr12_-_11150474 | 0.05 |
ENST00000538986.1 |
TAS2R20 |
taste receptor, type 2, member 20 |
| chr17_+_34391625 | 0.05 |
ENST00000004921.3 |
CCL18 |
chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) |
| chr19_+_55385682 | 0.04 |
ENST00000391726.3 |
FCAR |
Fc fragment of IgA, receptor for |
| chr7_-_105029812 | 0.04 |
ENST00000482897.1 |
SRPK2 |
SRSF protein kinase 2 |
| chr3_-_58613323 | 0.04 |
ENST00000474531.1 ENST00000465970.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr19_+_17830051 | 0.04 |
ENST00000594625.1 ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S |
microtubule-associated protein 1S |
| chr11_-_47400078 | 0.04 |
ENST00000378538.3 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr19_+_49588690 | 0.04 |
ENST00000221448.5 |
SNRNP70 |
small nuclear ribonucleoprotein 70kDa (U1) |
| chr1_-_52499443 | 0.04 |
ENST00000371614.1 |
KTI12 |
KTI12 homolog, chromatin associated (S. cerevisiae) |
| chr11_-_5255696 | 0.04 |
ENST00000292901.3 ENST00000417377.1 |
HBD |
hemoglobin, delta |
| chr2_+_171034646 | 0.04 |
ENST00000409044.3 ENST00000408978.4 |
MYO3B |
myosin IIIB |
| chr1_-_43638168 | 0.04 |
ENST00000431635.2 |
EBNA1BP2 |
EBNA1 binding protein 2 |
| chr22_-_30662828 | 0.04 |
ENST00000403463.1 ENST00000215781.2 |
OSM |
oncostatin M |
| chr10_-_73976025 | 0.03 |
ENST00000342444.4 ENST00000533958.1 ENST00000527593.1 ENST00000394915.3 ENST00000530461.1 ENST00000317168.6 ENST00000524829.1 |
ASCC1 |
activating signal cointegrator 1 complex subunit 1 |
| chr11_+_102980126 | 0.03 |
ENST00000375735.2 |
DYNC2H1 |
dynein, cytoplasmic 2, heavy chain 1 |
| chr12_-_110434096 | 0.03 |
ENST00000320063.9 ENST00000457474.2 ENST00000547815.1 ENST00000361006.5 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr2_+_171036635 | 0.03 |
ENST00000484338.2 ENST00000334231.6 |
MYO3B |
myosin IIIB |
| chr3_+_178276488 | 0.03 |
ENST00000432997.1 ENST00000455865.1 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr10_-_73976884 | 0.03 |
ENST00000317126.4 ENST00000545550.1 |
ASCC1 |
activating signal cointegrator 1 complex subunit 1 |
| chr15_+_59903975 | 0.03 |
ENST00000560585.1 ENST00000396065.1 |
GCNT3 |
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr10_-_43892668 | 0.03 |
ENST00000544000.1 |
HNRNPF |
heterogeneous nuclear ribonucleoprotein F |
| chr19_-_10491130 | 0.03 |
ENST00000530829.1 ENST00000529370.1 |
TYK2 |
tyrosine kinase 2 |
| chr4_+_154622652 | 0.03 |
ENST00000260010.6 |
TLR2 |
toll-like receptor 2 |
| chr11_-_104827425 | 0.03 |
ENST00000393150.3 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr19_+_21324827 | 0.03 |
ENST00000600692.1 ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431 |
zinc finger protein 431 |
| chr14_+_62462541 | 0.02 |
ENST00000430451.2 |
SYT16 |
synaptotagmin XVI |
| chr2_-_14541060 | 0.02 |
ENST00000418420.1 ENST00000417751.1 |
LINC00276 |
long intergenic non-protein coding RNA 276 |
| chr1_-_212004090 | 0.02 |
ENST00000366997.4 |
LPGAT1 |
lysophosphatidylglycerol acyltransferase 1 |
| chr12_+_100594557 | 0.02 |
ENST00000546902.1 ENST00000552376.1 ENST00000551617.1 |
ACTR6 |
ARP6 actin-related protein 6 homolog (yeast) |
| chr1_-_86848760 | 0.02 |
ENST00000460698.2 |
ODF2L |
outer dense fiber of sperm tails 2-like |
| chr3_-_126327398 | 0.02 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr11_+_125496124 | 0.02 |
ENST00000533778.2 ENST00000534070.1 |
CHEK1 |
checkpoint kinase 1 |
| chr11_-_113577014 | 0.02 |
ENST00000544634.1 ENST00000539732.1 ENST00000538770.1 ENST00000536856.1 ENST00000544476.1 |
TMPRSS5 |
transmembrane protease, serine 5 |
| chr18_-_51751132 | 0.02 |
ENST00000256429.3 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr11_-_26593677 | 0.02 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr18_+_21719018 | 0.02 |
ENST00000585037.1 ENST00000415309.2 ENST00000399481.2 ENST00000577705.1 ENST00000327201.6 |
CABYR |
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr17_+_75181292 | 0.02 |
ENST00000431431.2 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
| chr6_+_27833034 | 0.01 |
ENST00000357320.2 |
HIST1H2AL |
histone cluster 1, H2al |
| chr3_+_127770455 | 0.01 |
ENST00000464451.1 |
SEC61A1 |
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr19_+_49588677 | 0.01 |
ENST00000598984.1 ENST00000598441.1 |
SNRNP70 |
small nuclear ribonucleoprotein 70kDa (U1) |
| chr1_+_50459990 | 0.01 |
ENST00000448346.1 |
AL645730.2 |
AL645730.2 |
| chr19_+_3762703 | 0.01 |
ENST00000589174.1 |
MRPL54 |
mitochondrial ribosomal protein L54 |
| chr4_+_156680153 | 0.01 |
ENST00000502959.1 ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3 |
guanylate cyclase 1, soluble, beta 3 |
| chr11_+_125496400 | 0.01 |
ENST00000524737.1 |
CHEK1 |
checkpoint kinase 1 |
| chr1_+_65613217 | 0.01 |
ENST00000545314.1 |
AK4 |
adenylate kinase 4 |
| chr11_+_125496619 | 0.01 |
ENST00000532669.1 ENST00000278916.3 |
CHEK1 |
checkpoint kinase 1 |
| chr11_-_76381029 | 0.00 |
ENST00000407242.2 ENST00000421973.1 |
LRRC32 |
leucine rich repeat containing 32 |
| chr12_+_93096619 | 0.00 |
ENST00000397833.3 |
C12orf74 |
chromosome 12 open reading frame 74 |
| chr3_+_132316081 | 0.00 |
ENST00000249887.2 |
ACKR4 |
atypical chemokine receptor 4 |
| chr19_-_10491234 | 0.00 |
ENST00000524462.1 ENST00000531836.1 ENST00000525621.1 |
TYK2 |
tyrosine kinase 2 |
| chr6_-_46048116 | 0.00 |
ENST00000185206.6 |
CLIC5 |
chloride intracellular channel 5 |
| chr14_+_20811722 | 0.00 |
ENST00000429687.3 |
PARP2 |
poly (ADP-ribose) polymerase 2 |
| chr1_+_202789394 | 0.00 |
ENST00000330493.5 |
RP11-480I12.4 |
Putative inactive alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase-like protein LOC641515 |
| chr7_-_92848858 | 0.00 |
ENST00000440868.1 |
HEPACAM2 |
HEPACAM family member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 0.3 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.0 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |