Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
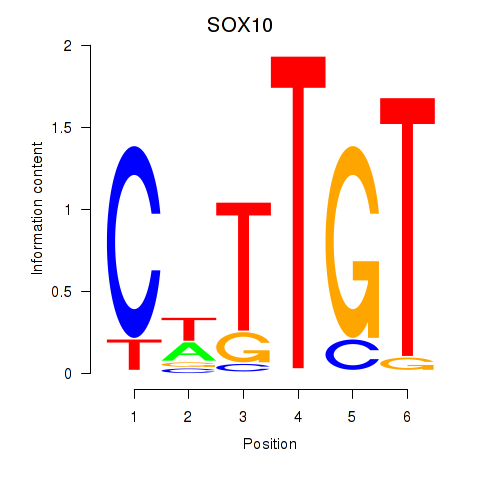
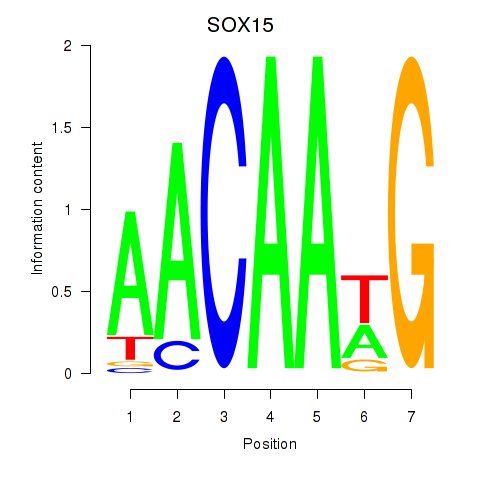
Results for SOX10_SOX15
Z-value: 1.14
Transcription factors associated with SOX10_SOX15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX10
|
ENSG00000100146.12 | SOX10 |
|
SOX15
|
ENSG00000129194.3 | SOX15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX15 | hg19_v2_chr17_-_7493390_7493488 | -0.90 | 2.2e-03 | Click! |
| SOX10 | hg19_v2_chr22_-_38380543_38380569 | -0.81 | 1.6e-02 | Click! |
Activity profile of SOX10_SOX15 motif
Sorted Z-values of SOX10_SOX15 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX10_SOX15
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_159141397 | 1.67 |
ENST00000368124.4 ENST00000368125.4 ENST00000416746.1 |
CADM3 |
cell adhesion molecule 3 |
| chr1_+_163038565 | 0.85 |
ENST00000421743.2 |
RGS4 |
regulator of G-protein signaling 4 |
| chr5_-_111093081 | 0.82 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr5_-_111092873 | 0.79 |
ENST00000509025.1 ENST00000515855.1 |
NREP |
neuronal regeneration related protein |
| chr5_-_111093167 | 0.70 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr5_-_111092930 | 0.67 |
ENST00000257435.7 |
NREP |
neuronal regeneration related protein |
| chr8_+_97597148 | 0.64 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr1_+_183774240 | 0.61 |
ENST00000360851.3 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr14_-_51027838 | 0.59 |
ENST00000555216.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr6_+_114178512 | 0.58 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr3_+_151986709 | 0.56 |
ENST00000495875.2 ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr15_+_63335899 | 0.53 |
ENST00000561266.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chrX_+_12993202 | 0.52 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr6_+_72596604 | 0.52 |
ENST00000348717.5 ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr1_+_215256467 | 0.49 |
ENST00000391894.2 ENST00000444842.2 |
KCNK2 |
potassium channel, subfamily K, member 2 |
| chrX_+_80457442 | 0.49 |
ENST00000373212.5 |
SH3BGRL |
SH3 domain binding glutamic acid-rich protein like |
| chr3_+_141105235 | 0.49 |
ENST00000503809.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr20_-_45985172 | 0.48 |
ENST00000536340.1 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr5_-_150521192 | 0.46 |
ENST00000523714.1 ENST00000521749.1 |
ANXA6 |
annexin A6 |
| chr15_-_52944231 | 0.45 |
ENST00000546305.2 |
FAM214A |
family with sequence similarity 214, member A |
| chr7_+_44788430 | 0.42 |
ENST00000457123.1 ENST00000309315.4 |
ZMIZ2 |
zinc finger, MIZ-type containing 2 |
| chr12_+_53443680 | 0.42 |
ENST00000314250.6 ENST00000451358.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr12_+_53443963 | 0.42 |
ENST00000546602.1 ENST00000552570.1 ENST00000549700.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr1_+_47799542 | 0.40 |
ENST00000471289.2 ENST00000450808.2 |
CMPK1 |
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chrX_-_142722897 | 0.38 |
ENST00000338017.4 |
SLITRK4 |
SLIT and NTRK-like family, member 4 |
| chr4_-_138453606 | 0.36 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr17_+_75447326 | 0.35 |
ENST00000591088.1 |
SEPT9 |
septin 9 |
| chrX_+_12993336 | 0.35 |
ENST00000380635.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr11_-_66445219 | 0.35 |
ENST00000525754.1 ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B |
RNA binding motif protein 4B |
| chr5_-_65017921 | 0.34 |
ENST00000381007.4 |
SGTB |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr3_+_133118839 | 0.33 |
ENST00000302334.2 |
BFSP2 |
beaded filament structural protein 2, phakinin |
| chrX_-_106960285 | 0.33 |
ENST00000503515.1 ENST00000372397.2 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr20_-_45984401 | 0.32 |
ENST00000311275.7 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr2_-_188312971 | 0.32 |
ENST00000410068.1 ENST00000447403.1 ENST00000410102.1 |
CALCRL |
calcitonin receptor-like |
| chr13_-_33780133 | 0.31 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr20_+_33292068 | 0.30 |
ENST00000374810.3 ENST00000374809.2 ENST00000451665.1 |
TP53INP2 |
tumor protein p53 inducible nuclear protein 2 |
| chr1_-_26233423 | 0.30 |
ENST00000357865.2 |
STMN1 |
stathmin 1 |
| chr7_-_47621736 | 0.30 |
ENST00000311160.9 |
TNS3 |
tensin 3 |
| chr16_-_73082274 | 0.29 |
ENST00000268489.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr2_-_37899323 | 0.29 |
ENST00000295324.3 ENST00000457889.1 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
| chr5_-_14871866 | 0.27 |
ENST00000284268.6 |
ANKH |
ANKH inorganic pyrophosphate transport regulator |
| chr6_+_108881012 | 0.27 |
ENST00000343882.6 |
FOXO3 |
forkhead box O3 |
| chr2_-_179343226 | 0.27 |
ENST00000434643.2 |
FKBP7 |
FK506 binding protein 7 |
| chr1_+_197886461 | 0.27 |
ENST00000367388.3 ENST00000337020.2 ENST00000367387.4 |
LHX9 |
LIM homeobox 9 |
| chr12_+_15475462 | 0.26 |
ENST00000543886.1 ENST00000348962.2 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr2_-_179343268 | 0.26 |
ENST00000424785.2 |
FKBP7 |
FK506 binding protein 7 |
| chr3_-_18466026 | 0.25 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr18_+_6729698 | 0.25 |
ENST00000383472.4 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr1_-_79472365 | 0.25 |
ENST00000370742.3 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr17_-_9940058 | 0.24 |
ENST00000585266.1 |
GAS7 |
growth arrest-specific 7 |
| chr15_-_82338460 | 0.24 |
ENST00000558133.1 ENST00000329713.4 |
MEX3B |
mex-3 RNA binding family member B |
| chr1_+_64239657 | 0.23 |
ENST00000371080.1 ENST00000371079.1 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
| chr2_-_166060571 | 0.23 |
ENST00000360093.3 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr6_+_56911476 | 0.22 |
ENST00000545356.1 |
KIAA1586 |
KIAA1586 |
| chr16_+_53242350 | 0.22 |
ENST00000565442.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr1_+_201857798 | 0.22 |
ENST00000362011.6 |
SHISA4 |
shisa family member 4 |
| chr13_-_41593425 | 0.21 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr9_-_120177342 | 0.21 |
ENST00000361209.2 |
ASTN2 |
astrotactin 2 |
| chr20_+_61448376 | 0.21 |
ENST00000343916.3 |
COL9A3 |
collagen, type IX, alpha 3 |
| chr18_+_72201664 | 0.21 |
ENST00000358821.3 |
CNDP1 |
carnosine dipeptidase 1 (metallopeptidase M20 family) |
| chr1_-_182360498 | 0.21 |
ENST00000417584.2 |
GLUL |
glutamate-ammonia ligase |
| chr16_-_30798492 | 0.20 |
ENST00000262525.4 |
ZNF629 |
zinc finger protein 629 |
| chr2_-_166060552 | 0.20 |
ENST00000283254.7 ENST00000453007.1 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr2_-_175711133 | 0.20 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr22_+_39052632 | 0.20 |
ENST00000411557.1 ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1 |
chibby homolog 1 (Drosophila) |
| chr11_+_20044600 | 0.20 |
ENST00000311043.8 |
NAV2 |
neuron navigator 2 |
| chr5_+_140729649 | 0.19 |
ENST00000523390.1 |
PCDHGB1 |
protocadherin gamma subfamily B, 1 |
| chr1_+_33722080 | 0.19 |
ENST00000483388.1 ENST00000539719.1 |
ZNF362 |
zinc finger protein 362 |
| chr2_+_120517174 | 0.18 |
ENST00000263708.2 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr20_-_30310797 | 0.17 |
ENST00000422920.1 |
BCL2L1 |
BCL2-like 1 |
| chr9_+_96846740 | 0.17 |
ENST00000288976.3 |
PTPDC1 |
protein tyrosine phosphatase domain containing 1 |
| chr9_-_73483958 | 0.17 |
ENST00000377101.1 ENST00000377106.1 ENST00000360823.2 ENST00000377105.1 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr11_-_111781454 | 0.17 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr5_+_138089100 | 0.17 |
ENST00000520339.1 ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr6_+_119215308 | 0.17 |
ENST00000229595.5 |
ASF1A |
anti-silencing function 1A histone chaperone |
| chr10_+_22605374 | 0.17 |
ENST00000448361.1 |
COMMD3 |
COMM domain containing 3 |
| chr1_-_11751529 | 0.17 |
ENST00000376672.1 |
MAD2L2 |
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr2_-_72374948 | 0.17 |
ENST00000546307.1 ENST00000474509.1 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr4_+_71588372 | 0.16 |
ENST00000536664.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr11_-_9286921 | 0.16 |
ENST00000328194.3 |
DENND5A |
DENN/MADD domain containing 5A |
| chr4_-_140477928 | 0.16 |
ENST00000274031.3 |
SETD7 |
SET domain containing (lysine methyltransferase) 7 |
| chr1_-_154155675 | 0.16 |
ENST00000330188.9 ENST00000341485.5 |
TPM3 |
tropomyosin 3 |
| chr15_+_52043758 | 0.16 |
ENST00000249700.4 ENST00000539962.2 |
TMOD2 |
tropomodulin 2 (neuronal) |
| chr12_+_104359641 | 0.16 |
ENST00000537100.1 |
TDG |
thymine-DNA glycosylase |
| chr10_-_99393208 | 0.16 |
ENST00000307450.6 |
MORN4 |
MORN repeat containing 4 |
| chr1_-_95391315 | 0.16 |
ENST00000545882.1 ENST00000415017.1 |
CNN3 |
calponin 3, acidic |
| chr18_+_6729725 | 0.16 |
ENST00000400091.2 ENST00000583410.1 ENST00000584387.1 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr8_-_124553437 | 0.15 |
ENST00000517956.1 ENST00000443022.2 |
FBXO32 |
F-box protein 32 |
| chr1_-_11751665 | 0.15 |
ENST00000376667.3 ENST00000235310.3 |
MAD2L2 |
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chrX_+_10124977 | 0.15 |
ENST00000380833.4 |
CLCN4 |
chloride channel, voltage-sensitive 4 |
| chrX_-_13956497 | 0.15 |
ENST00000398361.3 |
GPM6B |
glycoprotein M6B |
| chr12_-_42631529 | 0.15 |
ENST00000548917.1 |
YAF2 |
YY1 associated factor 2 |
| chr18_+_72201829 | 0.15 |
ENST00000582365.1 |
CNDP1 |
carnosine dipeptidase 1 (metallopeptidase M20 family) |
| chr11_-_111781610 | 0.15 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr16_+_53241854 | 0.15 |
ENST00000565803.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr11_-_111781554 | 0.15 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr14_+_77228532 | 0.15 |
ENST00000167106.4 ENST00000554237.1 |
VASH1 |
vasohibin 1 |
| chr4_+_150999418 | 0.15 |
ENST00000296550.7 |
DCLK2 |
doublecortin-like kinase 2 |
| chr15_-_57210769 | 0.15 |
ENST00000559000.1 |
ZNF280D |
zinc finger protein 280D |
| chr12_-_109915098 | 0.14 |
ENST00000542858.1 ENST00000542262.1 ENST00000424763.2 |
KCTD10 |
potassium channel tetramerization domain containing 10 |
| chr3_-_151102529 | 0.14 |
ENST00000302632.3 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
| chr7_-_27142290 | 0.14 |
ENST00000222718.5 |
HOXA2 |
homeobox A2 |
| chr12_+_32260085 | 0.14 |
ENST00000548411.1 ENST00000281474.5 ENST00000551086.1 |
BICD1 |
bicaudal D homolog 1 (Drosophila) |
| chr12_+_26111823 | 0.14 |
ENST00000381352.3 ENST00000535907.1 ENST00000405154.2 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr18_+_3448455 | 0.14 |
ENST00000549780.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chrX_+_70503433 | 0.14 |
ENST00000276079.8 ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO |
non-POU domain containing, octamer-binding |
| chr2_-_163099885 | 0.13 |
ENST00000443424.1 |
FAP |
fibroblast activation protein, alpha |
| chr5_+_140864649 | 0.13 |
ENST00000306593.1 |
PCDHGC4 |
protocadherin gamma subfamily C, 4 |
| chr1_-_113247543 | 0.13 |
ENST00000414971.1 ENST00000534717.1 |
RHOC |
ras homolog family member C |
| chr10_+_22605304 | 0.13 |
ENST00000475460.2 ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1 COMMD3 |
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr2_-_86564776 | 0.13 |
ENST00000165698.5 ENST00000541910.1 ENST00000535845.1 |
REEP1 |
receptor accessory protein 1 |
| chr9_-_14308004 | 0.13 |
ENST00000493697.1 |
NFIB |
nuclear factor I/B |
| chr5_+_140743859 | 0.13 |
ENST00000518069.1 |
PCDHGA5 |
protocadherin gamma subfamily A, 5 |
| chr5_-_171711061 | 0.13 |
ENST00000393792.2 |
UBTD2 |
ubiquitin domain containing 2 |
| chr7_-_128415844 | 0.13 |
ENST00000249389.2 |
OPN1SW |
opsin 1 (cone pigments), short-wave-sensitive |
| chr14_-_50999190 | 0.13 |
ENST00000557390.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr4_-_138453559 | 0.13 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr17_+_7461613 | 0.12 |
ENST00000438470.1 ENST00000436057.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr11_-_6677018 | 0.12 |
ENST00000299441.3 |
DCHS1 |
dachsous cadherin-related 1 |
| chr19_+_1249869 | 0.12 |
ENST00000591446.2 |
MIDN |
midnolin |
| chr13_+_30002846 | 0.12 |
ENST00000542829.1 |
MTUS2 |
microtubule associated tumor suppressor candidate 2 |
| chr11_+_111807863 | 0.12 |
ENST00000440460.2 |
DIXDC1 |
DIX domain containing 1 |
| chr2_-_85625857 | 0.12 |
ENST00000453973.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr1_+_228337553 | 0.12 |
ENST00000366714.2 |
GJC2 |
gap junction protein, gamma 2, 47kDa |
| chr5_+_43602750 | 0.12 |
ENST00000505678.2 ENST00000512422.1 ENST00000264663.5 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr8_+_81398444 | 0.11 |
ENST00000455036.3 ENST00000426744.2 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
| chr10_-_70092671 | 0.11 |
ENST00000358769.2 ENST00000432941.1 ENST00000495025.2 |
PBLD |
phenazine biosynthesis-like protein domain containing |
| chr20_+_11898507 | 0.11 |
ENST00000378226.2 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr7_-_111846435 | 0.11 |
ENST00000437633.1 ENST00000428084.1 |
DOCK4 |
dedicator of cytokinesis 4 |
| chr10_+_35484793 | 0.11 |
ENST00000488741.1 ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM |
cAMP responsive element modulator |
| chr3_-_46904946 | 0.11 |
ENST00000292327.4 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr3_-_46904918 | 0.11 |
ENST00000395869.1 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr15_+_57210961 | 0.11 |
ENST00000557843.1 |
TCF12 |
transcription factor 12 |
| chr1_-_151431909 | 0.11 |
ENST00000361398.3 ENST00000271715.2 |
POGZ |
pogo transposable element with ZNF domain |
| chr7_+_134551583 | 0.11 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr11_-_89224299 | 0.11 |
ENST00000343727.5 ENST00000531342.1 ENST00000375979.3 |
NOX4 |
NADPH oxidase 4 |
| chr16_+_30406721 | 0.11 |
ENST00000320159.2 |
ZNF48 |
zinc finger protein 48 |
| chr4_-_5890145 | 0.11 |
ENST00000397890.2 |
CRMP1 |
collapsin response mediator protein 1 |
| chr4_-_109090106 | 0.11 |
ENST00000379951.2 |
LEF1 |
lymphoid enhancer-binding factor 1 |
| chr17_-_56065484 | 0.11 |
ENST00000581208.1 |
VEZF1 |
vascular endothelial zinc finger 1 |
| chr10_-_70092635 | 0.10 |
ENST00000309049.4 |
PBLD |
phenazine biosynthesis-like protein domain containing |
| chr5_+_140710061 | 0.10 |
ENST00000517417.1 ENST00000378105.3 |
PCDHGA1 |
protocadherin gamma subfamily A, 1 |
| chr19_+_10765699 | 0.10 |
ENST00000590009.1 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
| chr16_-_15736881 | 0.10 |
ENST00000540441.2 |
KIAA0430 |
KIAA0430 |
| chr19_+_47105309 | 0.10 |
ENST00000599839.1 ENST00000596362.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr17_-_76713100 | 0.10 |
ENST00000585509.1 |
CYTH1 |
cytohesin 1 |
| chr19_-_5340730 | 0.10 |
ENST00000372412.4 ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
| chr22_-_41636929 | 0.10 |
ENST00000216241.9 |
CHADL |
chondroadherin-like |
| chr11_-_117166276 | 0.10 |
ENST00000510630.1 ENST00000392937.6 |
BACE1 |
beta-site APP-cleaving enzyme 1 |
| chr17_-_56605341 | 0.10 |
ENST00000583114.1 |
SEPT4 |
septin 4 |
| chr16_-_46865047 | 0.10 |
ENST00000394806.2 |
C16orf87 |
chromosome 16 open reading frame 87 |
| chr11_-_10830463 | 0.10 |
ENST00000527419.1 ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr3_-_71353892 | 0.10 |
ENST00000484350.1 |
FOXP1 |
forkhead box P1 |
| chr17_-_46690839 | 0.10 |
ENST00000498634.2 |
HOXB8 |
homeobox B8 |
| chr6_+_101846664 | 0.10 |
ENST00000421544.1 ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
| chr1_-_225840747 | 0.10 |
ENST00000366843.2 ENST00000366844.3 |
ENAH |
enabled homolog (Drosophila) |
| chr5_-_114515734 | 0.10 |
ENST00000514154.1 ENST00000282369.3 |
TRIM36 |
tripartite motif containing 36 |
| chr4_-_109089573 | 0.10 |
ENST00000265165.1 |
LEF1 |
lymphoid enhancer-binding factor 1 |
| chr11_+_126081662 | 0.09 |
ENST00000528985.1 ENST00000529731.1 ENST00000360194.4 ENST00000530043.1 |
FAM118B |
family with sequence similarity 118, member B |
| chr19_-_38720294 | 0.09 |
ENST00000412732.1 ENST00000456296.1 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
| chr19_-_18717627 | 0.09 |
ENST00000392386.3 |
CRLF1 |
cytokine receptor-like factor 1 |
| chr1_+_185703513 | 0.09 |
ENST00000271588.4 ENST00000367492.2 |
HMCN1 |
hemicentin 1 |
| chr3_-_48700310 | 0.09 |
ENST00000164024.4 ENST00000544264.1 |
CELSR3 |
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr10_+_35415719 | 0.09 |
ENST00000474362.1 ENST00000374721.3 |
CREM |
cAMP responsive element modulator |
| chr9_-_73736511 | 0.09 |
ENST00000377110.3 ENST00000377111.2 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr19_-_38720354 | 0.09 |
ENST00000416611.1 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
| chr1_+_93645314 | 0.09 |
ENST00000343253.7 |
CCDC18 |
coiled-coil domain containing 18 |
| chr2_+_149402553 | 0.09 |
ENST00000258484.6 ENST00000409654.1 |
EPC2 |
enhancer of polycomb homolog 2 (Drosophila) |
| chr17_-_53046058 | 0.09 |
ENST00000571584.1 ENST00000299335.3 |
COX11 |
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr17_+_75283973 | 0.09 |
ENST00000431235.2 ENST00000449803.2 |
SEPT9 |
septin 9 |
| chrX_-_21776281 | 0.09 |
ENST00000379494.3 |
SMPX |
small muscle protein, X-linked |
| chr15_+_92937058 | 0.09 |
ENST00000268164.3 |
ST8SIA2 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr1_+_244998602 | 0.09 |
ENST00000411948.2 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
| chr4_-_76598544 | 0.09 |
ENST00000515457.1 ENST00000357854.3 |
G3BP2 |
GTPase activating protein (SH3 domain) binding protein 2 |
| chr5_+_140480083 | 0.09 |
ENST00000231130.2 |
PCDHB3 |
protocadherin beta 3 |
| chr22_+_27053422 | 0.08 |
ENST00000413665.1 ENST00000421151.1 ENST00000456129.1 ENST00000430080.1 |
MIAT |
myocardial infarction associated transcript (non-protein coding) |
| chr10_+_35415978 | 0.08 |
ENST00000429130.3 ENST00000469949.2 ENST00000460270.1 |
CREM |
cAMP responsive element modulator |
| chr14_-_35099315 | 0.08 |
ENST00000396526.3 ENST00000396534.3 ENST00000355110.5 ENST00000557265.1 |
SNX6 |
sorting nexin 6 |
| chr22_-_36357671 | 0.08 |
ENST00000408983.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr22_+_19710468 | 0.08 |
ENST00000366425.3 |
GP1BB |
glycoprotein Ib (platelet), beta polypeptide |
| chr17_+_26698677 | 0.08 |
ENST00000457710.3 |
SARM1 |
sterile alpha and TIR motif containing 1 |
| chr13_+_49684445 | 0.08 |
ENST00000398316.3 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr8_+_81397876 | 0.08 |
ENST00000430430.1 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
| chr22_+_31742875 | 0.08 |
ENST00000504184.2 |
AC005003.1 |
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr10_+_99079008 | 0.08 |
ENST00000371021.3 |
FRAT1 |
frequently rearranged in advanced T-cell lymphomas |
| chr3_-_167813672 | 0.08 |
ENST00000470487.1 |
GOLIM4 |
golgi integral membrane protein 4 |
| chr6_+_64282447 | 0.08 |
ENST00000370650.2 ENST00000578299.1 |
PTP4A1 |
protein tyrosine phosphatase type IVA, member 1 |
| chr8_-_121824374 | 0.07 |
ENST00000517992.1 |
SNTB1 |
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr12_-_71551868 | 0.07 |
ENST00000247829.3 |
TSPAN8 |
tetraspanin 8 |
| chr15_+_62359175 | 0.07 |
ENST00000355522.5 |
C2CD4A |
C2 calcium-dependent domain containing 4A |
| chr14_-_91884150 | 0.07 |
ENST00000553403.1 |
CCDC88C |
coiled-coil domain containing 88C |
| chr5_+_43603229 | 0.07 |
ENST00000344920.4 ENST00000512996.2 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr2_+_74685527 | 0.07 |
ENST00000393972.3 ENST00000409737.1 ENST00000428943.1 |
WBP1 |
WW domain binding protein 1 |
| chr6_-_31632962 | 0.07 |
ENST00000456540.1 ENST00000445768.1 |
GPANK1 |
G patch domain and ankyrin repeats 1 |
| chr5_-_157002775 | 0.07 |
ENST00000257527.4 |
ADAM19 |
ADAM metallopeptidase domain 19 |
| chr4_+_76649797 | 0.07 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr14_-_71107921 | 0.07 |
ENST00000553982.1 ENST00000500016.1 |
CTD-2540L5.5 CTD-2540L5.6 |
CTD-2540L5.5 CTD-2540L5.6 |
| chr6_-_79944336 | 0.07 |
ENST00000344726.5 ENST00000275036.7 |
HMGN3 |
high mobility group nucleosomal binding domain 3 |
| chr6_+_27114861 | 0.07 |
ENST00000377459.1 |
HIST1H2AH |
histone cluster 1, H2ah |
| chr6_+_126240442 | 0.07 |
ENST00000448104.1 ENST00000438495.1 ENST00000444128.1 |
NCOA7 |
nuclear receptor coactivator 7 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.4 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.8 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.9 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.3 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.5 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.4 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.2 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.2 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.3 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.5 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.1 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.1 | GO:0061030 | axon target recognition(GO:0007412) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.0 | 0.1 | GO:0070837 | L-ascorbic acid transport(GO:0015882) dehydroascorbic acid transport(GO:0070837) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.1 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.5 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.0 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.1 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.0 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.3 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.0 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.5 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 1.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.2 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.2 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0033300 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) dehydroascorbic acid transporter activity(GO:0033300) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |