Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
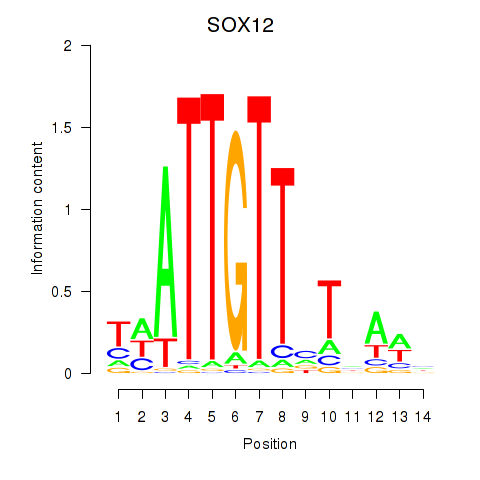
Results for SOX13_SOX12
Z-value: 0.63
Transcription factors associated with SOX13_SOX12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX13
|
ENSG00000143842.10 | SOX13 |
|
SOX12
|
ENSG00000177732.6 | SOX12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX12 | hg19_v2_chr20_+_306221_306239 | -0.54 | 1.6e-01 | Click! |
| SOX13 | hg19_v2_chr1_+_204042723_204042784 | 0.09 | 8.3e-01 | Click! |
Activity profile of SOX13_SOX12 motif
Sorted Z-values of SOX13_SOX12 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX13_SOX12
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_8803128 | 1.15 |
ENST00000543467.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr10_+_118187379 | 1.15 |
ENST00000369230.3 |
PNLIPRP3 |
pancreatic lipase-related protein 3 |
| chr1_+_153003671 | 1.13 |
ENST00000307098.4 |
SPRR1B |
small proline-rich protein 1B |
| chr12_-_8815215 | 0.93 |
ENST00000544889.1 ENST00000543369.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr12_-_8815299 | 0.78 |
ENST00000535336.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr19_+_35609380 | 0.74 |
ENST00000604621.1 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
| chr5_+_147582387 | 0.69 |
ENST00000325630.2 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr1_+_77333117 | 0.65 |
ENST00000477717.1 |
ST6GALNAC5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr1_-_153363452 | 0.63 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr10_-_105845674 | 0.58 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr6_-_136847610 | 0.57 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr5_+_140235469 | 0.56 |
ENST00000506939.2 ENST00000307360.5 |
PCDHA10 |
protocadherin alpha 10 |
| chr8_+_32579341 | 0.51 |
ENST00000519240.1 ENST00000539990.1 |
NRG1 |
neuregulin 1 |
| chr18_+_61254534 | 0.48 |
ENST00000269489.5 |
SERPINB13 |
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr12_-_8814669 | 0.47 |
ENST00000535411.1 ENST00000540087.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr2_+_207804278 | 0.47 |
ENST00000272852.3 |
CPO |
carboxypeptidase O |
| chr12_-_8815404 | 0.44 |
ENST00000359478.2 ENST00000396549.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr5_+_140213815 | 0.43 |
ENST00000525929.1 ENST00000378125.3 |
PCDHA7 |
protocadherin alpha 7 |
| chr3_+_98699880 | 0.41 |
ENST00000473756.1 |
LINC00973 |
long intergenic non-protein coding RNA 973 |
| chr11_-_51412448 | 0.41 |
ENST00000319760.6 |
OR4A5 |
olfactory receptor, family 4, subfamily A, member 5 |
| chr11_-_102651343 | 0.39 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr2_-_216878305 | 0.38 |
ENST00000263268.6 |
MREG |
melanoregulin |
| chr5_+_35856951 | 0.38 |
ENST00000303115.3 ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R |
interleukin 7 receptor |
| chr3_+_111717600 | 0.37 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr4_-_15939963 | 0.37 |
ENST00000259988.2 |
FGFBP1 |
fibroblast growth factor binding protein 1 |
| chr7_-_121944491 | 0.35 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr2_-_238499303 | 0.34 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr21_+_43619796 | 0.34 |
ENST00000398457.2 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr5_+_140248518 | 0.34 |
ENST00000398640.2 |
PCDHA11 |
protocadherin alpha 11 |
| chr2_+_102927962 | 0.33 |
ENST00000233954.1 ENST00000393393.3 ENST00000410040.1 |
IL1RL1 IL18R1 |
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr11_+_7110165 | 0.33 |
ENST00000306904.5 |
RBMXL2 |
RNA binding motif protein, X-linked-like 2 |
| chr3_+_111717511 | 0.33 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr18_+_61254570 | 0.33 |
ENST00000344731.5 |
SERPINB13 |
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr12_+_4385230 | 0.32 |
ENST00000536537.1 |
CCND2 |
cyclin D2 |
| chr1_+_161677034 | 0.32 |
ENST00000349527.4 ENST00000309691.6 ENST00000294796.4 ENST00000367953.3 ENST00000367950.1 |
FCRLA |
Fc receptor-like A |
| chr5_+_140220769 | 0.32 |
ENST00000531613.1 ENST00000378123.3 |
PCDHA8 |
protocadherin alpha 8 |
| chr1_+_152975488 | 0.31 |
ENST00000542696.1 |
SPRR3 |
small proline-rich protein 3 |
| chr5_+_140261703 | 0.31 |
ENST00000409494.1 ENST00000289272.2 |
PCDHA13 |
protocadherin alpha 13 |
| chr5_+_147582348 | 0.30 |
ENST00000514389.1 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr3_-_112564797 | 0.30 |
ENST00000398214.1 ENST00000448932.1 |
CD200R1L |
CD200 receptor 1-like |
| chr10_+_32873190 | 0.29 |
ENST00000375025.4 |
C10orf68 |
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr12_-_28125638 | 0.28 |
ENST00000545234.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr4_+_77356248 | 0.28 |
ENST00000296043.6 |
SHROOM3 |
shroom family member 3 |
| chr12_-_11287243 | 0.28 |
ENST00000539585.1 |
TAS2R30 |
taste receptor, type 2, member 30 |
| chr5_-_82969405 | 0.27 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr2_+_131769256 | 0.25 |
ENST00000355771.3 |
ARHGEF4 |
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr12_+_51317788 | 0.25 |
ENST00000550502.1 |
METTL7A |
methyltransferase like 7A |
| chr6_-_136788001 | 0.25 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chr4_+_53728457 | 0.25 |
ENST00000248706.3 |
RASL11B |
RAS-like, family 11, member B |
| chr11_+_128563652 | 0.24 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr12_+_9102632 | 0.24 |
ENST00000539240.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr1_-_153433120 | 0.24 |
ENST00000368723.3 |
S100A7 |
S100 calcium binding protein A7 |
| chr9_-_39239171 | 0.23 |
ENST00000358144.2 |
CNTNAP3 |
contactin associated protein-like 3 |
| chr2_-_238499131 | 0.23 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr7_+_142364193 | 0.23 |
ENST00000390397.2 |
TRBV24-1 |
T cell receptor beta variable 24-1 |
| chr7_+_116660246 | 0.23 |
ENST00000434836.1 ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7 |
suppression of tumorigenicity 7 |
| chr6_-_11779014 | 0.23 |
ENST00000229583.5 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr9_-_140196703 | 0.22 |
ENST00000356628.2 |
NRARP |
NOTCH-regulated ankyrin repeat protein |
| chr18_+_47088401 | 0.22 |
ENST00000261292.4 ENST00000427224.2 ENST00000580036.1 |
LIPG |
lipase, endothelial |
| chr3_+_32433154 | 0.22 |
ENST00000334983.5 ENST00000349718.4 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
| chr2_-_238499337 | 0.22 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr12_-_11150474 | 0.22 |
ENST00000538986.1 |
TAS2R20 |
taste receptor, type 2, member 20 |
| chr3_+_32433363 | 0.22 |
ENST00000465248.1 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
| chr1_+_161676739 | 0.21 |
ENST00000236938.6 ENST00000367959.2 ENST00000546024.1 ENST00000540521.1 ENST00000367949.2 ENST00000350710.3 ENST00000540926.1 |
FCRLA |
Fc receptor-like A |
| chr1_+_161676983 | 0.21 |
ENST00000367957.2 |
FCRLA |
Fc receptor-like A |
| chr6_+_106534192 | 0.21 |
ENST00000369091.2 ENST00000369096.4 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr14_+_22739823 | 0.21 |
ENST00000390464.2 |
TRAV38-1 |
T cell receptor alpha variable 38-1 |
| chr5_+_121465207 | 0.21 |
ENST00000296600.4 |
ZNF474 |
zinc finger protein 474 |
| chr4_+_100737954 | 0.21 |
ENST00000296414.7 ENST00000512369.1 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr11_+_128563948 | 0.21 |
ENST00000534087.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr8_+_102504651 | 0.20 |
ENST00000251808.3 ENST00000521085.1 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
| chr6_-_139613269 | 0.20 |
ENST00000358430.3 |
TXLNB |
taxilin beta |
| chr19_-_14785674 | 0.20 |
ENST00000253673.5 |
EMR3 |
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr14_+_22580233 | 0.20 |
ENST00000390454.2 |
TRAV25 |
T cell receptor alpha variable 25 |
| chr19_-_14785622 | 0.20 |
ENST00000443157.2 |
EMR3 |
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chrX_+_107288239 | 0.20 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr6_+_125540951 | 0.20 |
ENST00000524679.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr18_+_42260861 | 0.19 |
ENST00000282030.5 |
SETBP1 |
SET binding protein 1 |
| chr7_-_41742697 | 0.19 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr17_+_56315787 | 0.19 |
ENST00000262290.4 ENST00000421678.2 |
LPO |
lactoperoxidase |
| chr11_+_34654011 | 0.19 |
ENST00000531794.1 |
EHF |
ets homologous factor |
| chrX_+_107288197 | 0.19 |
ENST00000415430.3 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr19_-_14785698 | 0.18 |
ENST00000344373.4 ENST00000595472.1 |
EMR3 |
egf-like module containing, mucin-like, hormone receptor-like 3 |
| chr1_+_13910194 | 0.18 |
ENST00000376057.4 ENST00000510906.1 |
PDPN |
podoplanin |
| chr1_-_116383322 | 0.18 |
ENST00000429731.1 |
NHLH2 |
nescient helix loop helix 2 |
| chr12_-_10962767 | 0.18 |
ENST00000240691.2 |
TAS2R9 |
taste receptor, type 2, member 9 |
| chr6_+_12290586 | 0.18 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr6_-_11779174 | 0.18 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr19_-_43099070 | 0.18 |
ENST00000244336.5 |
CEACAM8 |
carcinoembryonic antigen-related cell adhesion molecule 8 |
| chr4_-_74864386 | 0.18 |
ENST00000296027.4 |
CXCL5 |
chemokine (C-X-C motif) ligand 5 |
| chr10_-_61513146 | 0.17 |
ENST00000430431.1 |
LINC00948 |
long intergenic non-protein coding RNA 948 |
| chr1_-_152297679 | 0.17 |
ENST00000368799.1 |
FLG |
filaggrin |
| chr1_-_153013588 | 0.17 |
ENST00000360379.3 |
SPRR2D |
small proline-rich protein 2D |
| chr9_+_22446814 | 0.17 |
ENST00000325870.2 |
DMRTA1 |
DMRT-like family A1 |
| chr2_+_90153696 | 0.17 |
ENST00000417279.2 |
IGKV3D-15 |
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr10_-_123357910 | 0.16 |
ENST00000336553.6 ENST00000457416.2 ENST00000360144.3 ENST00000369059.1 ENST00000356226.4 ENST00000351936.6 |
FGFR2 |
fibroblast growth factor receptor 2 |
| chr4_+_74301880 | 0.16 |
ENST00000395792.2 ENST00000226359.2 |
AFP |
alpha-fetoprotein |
| chr2_-_201599892 | 0.16 |
ENST00000452787.1 |
AC007163.3 |
AC007163.3 |
| chr12_+_25205446 | 0.16 |
ENST00000557489.1 ENST00000354454.3 ENST00000536173.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr10_-_116444371 | 0.16 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr11_-_79151695 | 0.16 |
ENST00000278550.7 |
TENM4 |
teneurin transmembrane protein 4 |
| chr10_+_5238793 | 0.16 |
ENST00000263126.1 |
AKR1C4 |
aldo-keto reductase family 1, member C4 |
| chr15_-_99791413 | 0.16 |
ENST00000394129.2 ENST00000558663.1 ENST00000394135.3 ENST00000561365.1 ENST00000560279.1 |
TTC23 |
tetratricopeptide repeat domain 23 |
| chr19_-_42192096 | 0.16 |
ENST00000602225.1 |
CEACAM7 |
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr4_+_187148556 | 0.15 |
ENST00000264690.6 ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1 |
kallikrein B, plasma (Fletcher factor) 1 |
| chr12_+_21168630 | 0.15 |
ENST00000421593.2 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr21_-_34186006 | 0.15 |
ENST00000490358.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr11_-_59950486 | 0.15 |
ENST00000426738.2 ENST00000533023.1 ENST00000420732.2 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr17_+_56315936 | 0.15 |
ENST00000543544.1 |
LPO |
lactoperoxidase |
| chr4_-_143226979 | 0.15 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr19_+_4402659 | 0.15 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chr18_-_70211691 | 0.15 |
ENST00000269503.4 |
CBLN2 |
cerebellin 2 precursor |
| chr11_+_55029628 | 0.14 |
ENST00000417545.2 |
TRIM48 |
tripartite motif containing 48 |
| chr4_-_143227088 | 0.14 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr7_-_36406750 | 0.14 |
ENST00000453212.1 ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895 |
KIAA0895 |
| chr16_+_2867228 | 0.14 |
ENST00000005995.3 ENST00000574813.1 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr19_+_41281282 | 0.14 |
ENST00000263369.3 |
MIA |
melanoma inhibitory activity |
| chr8_-_124054587 | 0.14 |
ENST00000259512.4 |
DERL1 |
derlin 1 |
| chr10_-_10836919 | 0.14 |
ENST00000602763.1 ENST00000415590.2 ENST00000434919.2 |
SFTA1P |
surfactant associated 1, pseudogene |
| chr6_-_155776966 | 0.14 |
ENST00000159060.2 |
NOX3 |
NADPH oxidase 3 |
| chr4_-_77328458 | 0.14 |
ENST00000388914.3 ENST00000434846.2 |
CCDC158 |
coiled-coil domain containing 158 |
| chr20_-_5426332 | 0.13 |
ENST00000420529.1 |
LINC00658 |
long intergenic non-protein coding RNA 658 |
| chr13_-_46716969 | 0.13 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr15_-_34610962 | 0.13 |
ENST00000290209.5 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr2_+_90198535 | 0.13 |
ENST00000390276.2 |
IGKV1D-12 |
immunoglobulin kappa variable 1D-12 |
| chr7_-_105162652 | 0.13 |
ENST00000356362.2 ENST00000469408.1 |
PUS7 |
pseudouridylate synthase 7 homolog (S. cerevisiae) |
| chr7_-_82792215 | 0.12 |
ENST00000333891.9 ENST00000423517.2 |
PCLO |
piccolo presynaptic cytomatrix protein |
| chrX_-_32173579 | 0.12 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr12_+_101988774 | 0.12 |
ENST00000545503.2 ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1 |
myosin binding protein C, slow type |
| chr1_+_153388993 | 0.12 |
ENST00000368729.4 |
S100A7A |
S100 calcium binding protein A7A |
| chr17_+_53828333 | 0.12 |
ENST00000268896.5 |
PCTP |
phosphatidylcholine transfer protein |
| chr15_-_76005170 | 0.12 |
ENST00000308508.5 |
CSPG4 |
chondroitin sulfate proteoglycan 4 |
| chr3_-_32544900 | 0.12 |
ENST00000205636.3 |
CMTM6 |
CKLF-like MARVEL transmembrane domain containing 6 |
| chr15_+_88120158 | 0.12 |
ENST00000560153.1 |
LINC00052 |
long intergenic non-protein coding RNA 52 |
| chr6_-_9939552 | 0.12 |
ENST00000460363.2 |
OFCC1 |
orofacial cleft 1 candidate 1 |
| chr1_-_153029980 | 0.12 |
ENST00000392653.2 |
SPRR2A |
small proline-rich protein 2A |
| chr21_-_34185989 | 0.12 |
ENST00000487113.1 ENST00000382373.4 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr14_+_50291993 | 0.12 |
ENST00000595378.1 |
AL627171.2 |
HCG1786899; PRO2610; Uncharacterized protein |
| chr11_-_8964580 | 0.11 |
ENST00000325884.1 |
ASCL3 |
achaete-scute family bHLH transcription factor 3 |
| chr6_+_150920999 | 0.11 |
ENST00000367328.1 ENST00000367326.1 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr2_+_197577841 | 0.11 |
ENST00000409270.1 |
CCDC150 |
coiled-coil domain containing 150 |
| chr14_-_106330458 | 0.11 |
ENST00000461719.1 |
IGHJ4 |
immunoglobulin heavy joining 4 |
| chr2_-_9143786 | 0.11 |
ENST00000462696.1 ENST00000305997.3 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
| chr12_+_25205666 | 0.11 |
ENST00000547044.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr10_+_49917751 | 0.11 |
ENST00000325239.5 ENST00000413659.2 |
WDFY4 |
WDFY family member 4 |
| chr9_-_21351377 | 0.11 |
ENST00000380210.1 |
IFNA6 |
interferon, alpha 6 |
| chr11_+_93479588 | 0.11 |
ENST00000526335.1 |
C11orf54 |
chromosome 11 open reading frame 54 |
| chr6_-_119256311 | 0.11 |
ENST00000316316.6 |
MCM9 |
minichromosome maintenance complex component 9 |
| chr16_+_2867164 | 0.11 |
ENST00000455114.1 ENST00000450020.3 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr11_+_73661364 | 0.11 |
ENST00000339764.1 |
DNAJB13 |
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr13_+_32313658 | 0.11 |
ENST00000380314.1 ENST00000298386.2 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
| chr15_+_80733570 | 0.10 |
ENST00000533983.1 ENST00000527771.1 ENST00000525103.1 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr11_-_89653576 | 0.10 |
ENST00000420869.1 |
TRIM49D1 |
tripartite motif containing 49D1 |
| chr11_-_10829851 | 0.10 |
ENST00000532082.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr7_+_99195677 | 0.10 |
ENST00000431679.1 |
GS1-259H13.2 |
GS1-259H13.2 |
| chr1_-_36107445 | 0.10 |
ENST00000373237.3 |
PSMB2 |
proteasome (prosome, macropain) subunit, beta type, 2 |
| chr10_-_61513201 | 0.10 |
ENST00000414264.1 ENST00000594536.1 |
LINC00948 |
long intergenic non-protein coding RNA 948 |
| chr2_+_118572226 | 0.10 |
ENST00000263239.2 |
DDX18 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr4_+_95128996 | 0.10 |
ENST00000457823.2 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr19_+_42212501 | 0.10 |
ENST00000398599.4 |
CEACAM5 |
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr18_+_55888767 | 0.10 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr20_-_50384864 | 0.10 |
ENST00000311637.5 ENST00000402822.1 |
ATP9A |
ATPase, class II, type 9A |
| chr12_-_10151773 | 0.10 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr19_+_41281060 | 0.10 |
ENST00000594436.1 ENST00000597784.1 |
MIA |
melanoma inhibitory activity |
| chr17_+_35851570 | 0.10 |
ENST00000394386.1 |
DUSP14 |
dual specificity phosphatase 14 |
| chr3_-_123339343 | 0.10 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr7_-_38305279 | 0.10 |
ENST00000443402.2 |
TRGC1 |
T cell receptor gamma constant 1 |
| chr21_+_25801041 | 0.10 |
ENST00000453784.2 ENST00000423581.1 |
AP000476.1 |
AP000476.1 |
| chr14_+_21387508 | 0.10 |
ENST00000555624.1 |
RP11-84C10.2 |
RP11-84C10.2 |
| chr6_-_28411241 | 0.10 |
ENST00000289788.4 |
ZSCAN23 |
zinc finger and SCAN domain containing 23 |
| chr3_+_47866490 | 0.10 |
ENST00000457607.1 |
DHX30 |
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr18_-_52989217 | 0.10 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr11_-_104817919 | 0.09 |
ENST00000533252.1 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr11_+_59480899 | 0.09 |
ENST00000300150.7 |
STX3 |
syntaxin 3 |
| chr12_-_31479045 | 0.09 |
ENST00000539409.1 ENST00000395766.1 |
FAM60A |
family with sequence similarity 60, member A |
| chr12_-_50616382 | 0.09 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr6_-_116381918 | 0.09 |
ENST00000606080.1 |
FRK |
fyn-related kinase |
| chr14_+_56584414 | 0.09 |
ENST00000559044.1 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
| chr11_+_33563821 | 0.09 |
ENST00000321505.4 ENST00000265654.5 ENST00000389726.3 |
KIAA1549L |
KIAA1549-like |
| chr12_+_20968608 | 0.09 |
ENST00000381541.3 ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3 SLCO1B3 SLCO1B7 |
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr11_-_59950519 | 0.09 |
ENST00000528851.1 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr12_+_48876275 | 0.09 |
ENST00000314014.2 |
C12orf54 |
chromosome 12 open reading frame 54 |
| chr14_+_73563735 | 0.09 |
ENST00000532192.1 |
RBM25 |
RNA binding motif protein 25 |
| chrX_-_15619076 | 0.09 |
ENST00000252519.3 |
ACE2 |
angiotensin I converting enzyme 2 |
| chr5_+_44809027 | 0.09 |
ENST00000507110.1 |
MRPS30 |
mitochondrial ribosomal protein S30 |
| chr12_+_40787194 | 0.09 |
ENST00000425730.2 ENST00000454784.4 |
MUC19 |
mucin 19, oligomeric |
| chr6_-_25830785 | 0.09 |
ENST00000468082.1 |
SLC17A1 |
solute carrier family 17 (organic anion transporter), member 1 |
| chrX_-_33229636 | 0.09 |
ENST00000357033.4 |
DMD |
dystrophin |
| chr4_+_154622652 | 0.09 |
ENST00000260010.6 |
TLR2 |
toll-like receptor 2 |
| chr1_-_158656488 | 0.09 |
ENST00000368147.4 |
SPTA1 |
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr1_+_87012753 | 0.09 |
ENST00000370563.3 |
CLCA4 |
chloride channel accessory 4 |
| chr8_+_104831472 | 0.09 |
ENST00000262231.10 ENST00000507740.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr3_+_171561127 | 0.09 |
ENST00000334567.5 ENST00000450693.1 |
TMEM212 |
transmembrane protein 212 |
| chr4_+_95128748 | 0.09 |
ENST00000359052.4 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chrX_+_105192423 | 0.08 |
ENST00000540278.1 |
NRK |
Nik related kinase |
| chr7_+_107220660 | 0.08 |
ENST00000465919.1 ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr1_-_225616515 | 0.08 |
ENST00000338179.2 ENST00000425080.1 |
LBR |
lamin B receptor |
| chr2_+_168725458 | 0.08 |
ENST00000392690.3 |
B3GALT1 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr15_-_79383102 | 0.08 |
ENST00000558480.2 ENST00000419573.3 |
RASGRF1 |
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr4_+_100432161 | 0.08 |
ENST00000326581.4 ENST00000514652.1 |
C4orf17 |
chromosome 4 open reading frame 17 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.4 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.3 | GO:0055099 | detection of endogenous stimulus(GO:0009726) response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 3.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.5 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.8 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 0.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.2 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.2 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.2 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.2 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.1 | 0.7 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.1 | 0.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.2 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.0 | 0.2 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.5 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.2 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 1.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0060720 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.2 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.0 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 1.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.2 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.0 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.3 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0060541 | lung development(GO:0030324) respiratory system development(GO:0060541) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.0 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.1 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.0 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.0 | GO:0098736 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.1 | GO:0035407 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.0 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.0 | 0.0 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 1.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.0 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.0 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.3 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.2 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 1.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 3.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.2 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) |
| 0.0 | 0.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |