Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
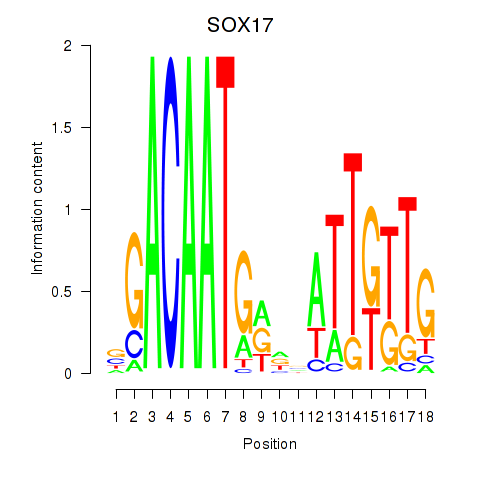
Results for SOX17
Z-value: 1.17
Transcription factors associated with SOX17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX17
|
ENSG00000164736.5 | SOX17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX17 | hg19_v2_chr8_+_55370487_55370503 | -0.45 | 2.7e-01 | Click! |
Activity profile of SOX17 motif
Sorted Z-values of SOX17 motif
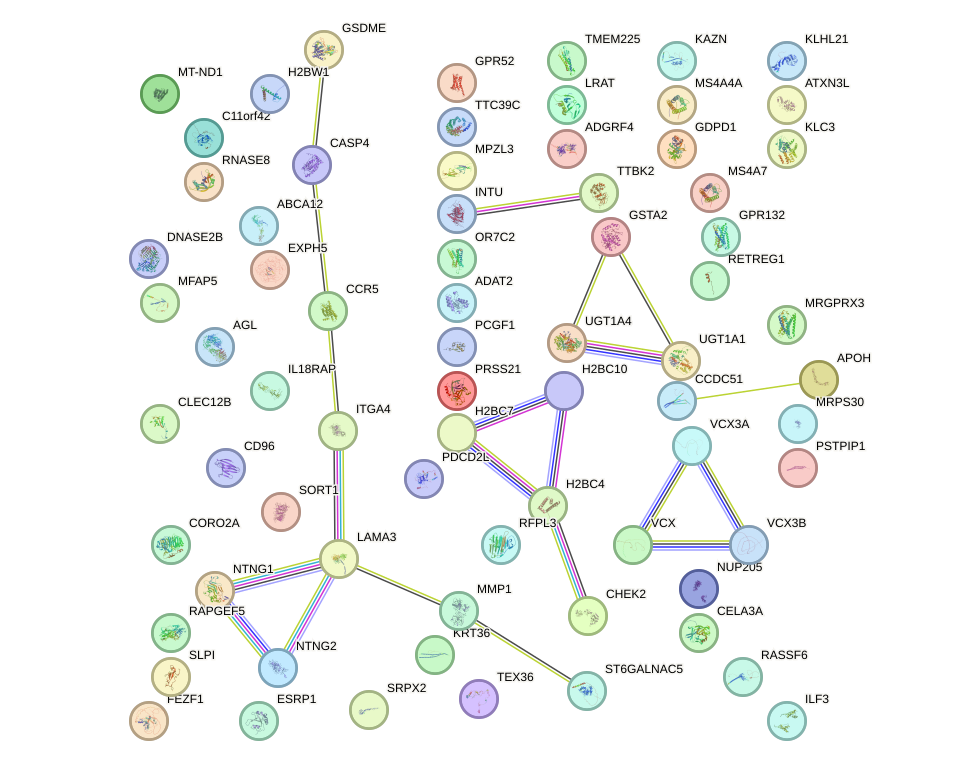
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX17
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_209602771 | 1.73 |
ENST00000440276.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr12_-_8815215 | 1.41 |
ENST00000544889.1 ENST00000543369.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr10_-_10836919 | 1.12 |
ENST00000602763.1 ENST00000415590.2 ENST00000434919.2 |
SFTA1P |
surfactant associated 1, pseudogene |
| chr10_-_10836865 | 1.11 |
ENST00000446372.2 |
SFTA1P |
surfactant associated 1, pseudogene |
| chr12_-_8815299 | 1.02 |
ENST00000535336.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr8_+_95653427 | 0.98 |
ENST00000454170.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr4_-_15939963 | 0.96 |
ENST00000259988.2 |
FGFBP1 |
fibroblast growth factor binding protein 1 |
| chr12_-_8814669 | 0.95 |
ENST00000535411.1 ENST00000540087.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr18_+_21452804 | 0.85 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr11_-_108464465 | 0.82 |
ENST00000525344.1 |
EXPH5 |
exophilin 5 |
| chr1_+_15256230 | 0.71 |
ENST00000376028.4 ENST00000400798.2 |
KAZN |
kazrin, periplakin interacting protein |
| chr7_-_22234381 | 0.70 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr11_-_118122996 | 0.69 |
ENST00000525386.1 ENST00000527472.1 ENST00000278949.4 |
MPZL3 |
myelin protein zero-like 3 |
| chr2_-_216003127 | 0.68 |
ENST00000412081.1 ENST00000272895.7 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr11_+_18154059 | 0.65 |
ENST00000531264.1 |
MRGPRX3 |
MAS-related GPR, member X3 |
| chr19_+_45843994 | 0.61 |
ENST00000391946.2 |
KLC3 |
kinesin light chain 3 |
| chr2_+_182321925 | 0.56 |
ENST00000339307.4 ENST00000397033.2 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
| chr14_+_21525981 | 0.52 |
ENST00000308227.2 |
RNASE8 |
ribonuclease, RNase A family, 8 |
| chr19_+_15052301 | 0.50 |
ENST00000248072.3 |
OR7C2 |
olfactory receptor, family 7, subfamily C, member 2 |
| chr19_+_35606777 | 0.49 |
ENST00000604404.1 ENST00000435734.2 ENST00000603181.1 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
| chr12_-_8815404 | 0.48 |
ENST00000359478.2 ENST00000396549.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr19_+_45844018 | 0.46 |
ENST00000585434.1 |
KLC3 |
kinesin light chain 3 |
| chr20_-_43883197 | 0.44 |
ENST00000338380.2 |
SLPI |
secretory leukocyte peptidase inhibitor |
| chr4_-_74486217 | 0.43 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr16_+_2867228 | 0.42 |
ENST00000005995.3 ENST00000574813.1 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr5_-_16509101 | 0.39 |
ENST00000399793.2 |
FAM134B |
family with sequence similarity 134, member B |
| chr1_+_77333117 | 0.38 |
ENST00000477717.1 |
ST6GALNAC5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr11_-_7695437 | 0.38 |
ENST00000533558.1 ENST00000527542.1 ENST00000531096.1 |
CYB5R2 |
cytochrome b5 reductase 2 |
| chr11_-_102668879 | 0.36 |
ENST00000315274.6 |
MMP1 |
matrix metallopeptidase 1 (interstitial collagenase) |
| chr12_+_10163231 | 0.35 |
ENST00000396502.1 ENST00000338896.5 |
CLEC12B |
C-type lectin domain family 12, member B |
| chr19_+_45844032 | 0.34 |
ENST00000589837.1 |
KLC3 |
kinesin light chain 3 |
| chr12_-_8815477 | 0.34 |
ENST00000433590.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr6_+_1080164 | 0.33 |
ENST00000314040.1 |
AL033381.1 |
Uncharacterized protein; cDNA FLJ34594 fis, clone KIDNE2009109 |
| chr4_+_155665123 | 0.31 |
ENST00000336356.3 |
LRAT |
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) |
| chr20_-_1309809 | 0.31 |
ENST00000360779.3 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
| chrX_-_13338518 | 0.30 |
ENST00000380622.2 |
ATXN3L |
ataxin 3-like |
| chr17_-_39646116 | 0.28 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr18_+_21269404 | 0.27 |
ENST00000313654.9 |
LAMA3 |
laminin, alpha 3 |
| chr22_-_29107919 | 0.26 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr1_+_107682629 | 0.26 |
ENST00000370074.4 ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1 |
netrin G1 |
| chr10_+_6622345 | 0.26 |
ENST00000445427.1 ENST00000455810.1 |
PRKCQ-AS1 |
PRKCQ antisense RNA 1 |
| chr19_+_6135646 | 0.26 |
ENST00000588304.1 ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2 |
acyl-CoA synthetase bubblegum family member 2 |
| chr11_+_6226782 | 0.26 |
ENST00000316375.2 |
C11orf42 |
chromosome 11 open reading frame 42 |
| chr8_+_21916710 | 0.25 |
ENST00000523266.1 ENST00000519907.1 |
DMTN |
dematin actin binding protein |
| chr10_-_135379132 | 0.22 |
ENST00000343131.5 |
SYCE1 |
synaptonemal complex central element protein 1 |
| chr4_-_87028478 | 0.22 |
ENST00000515400.1 ENST00000395157.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr5_+_44809027 | 0.22 |
ENST00000507110.1 |
MRPS30 |
mitochondrial ribosomal protein S30 |
| chr8_+_21155717 | 0.21 |
ENST00000522755.1 ENST00000519996.1 |
RP11-24P4.1 |
RP11-24P4.1 |
| chr15_+_88120158 | 0.21 |
ENST00000560153.1 |
LINC00052 |
long intergenic non-protein coding RNA 52 |
| chr11_-_104817919 | 0.21 |
ENST00000533252.1 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr11_-_123756334 | 0.20 |
ENST00000528595.1 ENST00000375026.2 |
TMEM225 |
transmembrane protein 225 |
| chr14_+_94577074 | 0.20 |
ENST00000444961.1 ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27 |
interferon, alpha-inducible protein 27 |
| chr4_-_74486347 | 0.20 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr16_+_2867164 | 0.20 |
ENST00000455114.1 ENST00000450020.3 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr6_-_52628271 | 0.19 |
ENST00000493422.1 |
GSTA2 |
glutathione S-transferase alpha 2 |
| chr1_+_22333943 | 0.19 |
ENST00000400271.2 |
CELA3A |
chymotrypsin-like elastase family, member 3A |
| chrX_-_103268259 | 0.19 |
ENST00000217926.5 |
H2BFWT |
H2B histone family, member W, testis-specific |
| chr6_-_19804973 | 0.19 |
ENST00000457670.1 ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2 |
RP4-625H18.2 |
| chr9_-_100935043 | 0.19 |
ENST00000343933.5 |
CORO2A |
coronin, actin binding protein, 2A |
| chr6_-_143771799 | 0.18 |
ENST00000237283.8 |
ADAT2 |
adenosine deaminase, tRNA-specific 2 |
| chr1_-_109935819 | 0.18 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr7_-_24797546 | 0.18 |
ENST00000414428.1 ENST00000419307.1 ENST00000342947.3 |
DFNA5 |
deafness, autosomal dominant 5 |
| chr1_-_205744205 | 0.18 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr18_+_21572737 | 0.17 |
ENST00000304621.6 |
TTC39C |
tetratricopeptide repeat domain 39C |
| chr15_+_77287426 | 0.17 |
ENST00000558012.1 ENST00000267939.5 ENST00000379595.3 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
| chr2_+_86668464 | 0.17 |
ENST00000409064.1 |
KDM3A |
lysine (K)-specific demethylase 3A |
| chr3_-_48481434 | 0.17 |
ENST00000395694.2 ENST00000447018.1 ENST00000442740.1 |
CCDC51 |
coiled-coil domain containing 51 |
| chr3_+_98451093 | 0.16 |
ENST00000483910.1 ENST00000460774.1 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr1_-_6662919 | 0.16 |
ENST00000377658.4 ENST00000377663.3 |
KLHL21 |
kelch-like family member 21 |
| chr6_+_47666275 | 0.16 |
ENST00000327753.3 ENST00000283303.2 |
GPR115 |
G protein-coupled receptor 115 |
| chr7_-_121944491 | 0.16 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr10_-_127371675 | 0.16 |
ENST00000532135.1 ENST00000526819.1 ENST00000368821.3 |
TEX36 |
testis expressed 36 |
| chr14_-_55369525 | 0.15 |
ENST00000543643.2 ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1 |
GTP cyclohydrolase 1 |
| chr19_+_10764937 | 0.15 |
ENST00000449870.1 ENST00000318511.3 ENST00000420083.1 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
| chr3_+_125687987 | 0.15 |
ENST00000514116.1 ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B |
rhophilin associated tail protein 1B |
| chr2_-_89278535 | 0.15 |
ENST00000390247.2 |
IGKV3-7 |
immunoglobulin kappa variable 3-7 (non-functional) |
| chr7_+_135242652 | 0.15 |
ENST00000285968.6 ENST00000440390.2 |
NUP205 |
nucleoporin 205kDa |
| chr14_-_105531759 | 0.15 |
ENST00000329797.3 ENST00000539291.2 ENST00000392585.2 |
GPR132 |
G protein-coupled receptor 132 |
| chrX_+_8432871 | 0.15 |
ENST00000381032.1 ENST00000453306.1 ENST00000444481.1 |
VCX3B |
variable charge, X-linked 3B |
| chr3_+_46412345 | 0.15 |
ENST00000292303.4 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr22_+_32753854 | 0.15 |
ENST00000249007.4 |
RFPL3 |
ret finger protein-like 3 |
| chr17_+_57297807 | 0.14 |
ENST00000284116.4 ENST00000581140.1 ENST00000581276.1 |
GDPD1 |
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr3_+_97887544 | 0.14 |
ENST00000356526.2 |
OR5H15 |
olfactory receptor, family 5, subfamily H, member 15 |
| chr17_-_64225508 | 0.14 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr22_+_32754139 | 0.14 |
ENST00000382088.3 |
RFPL3 |
ret finger protein-like 3 |
| chr2_+_103035102 | 0.14 |
ENST00000264260.2 |
IL18RAP |
interleukin 18 receptor accessory protein |
| chr14_+_39703112 | 0.14 |
ENST00000555143.1 ENST00000280082.3 |
MIA2 |
melanoma inhibitory activity 2 |
| chr2_-_74735707 | 0.14 |
ENST00000233630.6 |
PCGF1 |
polycomb group ring finger 1 |
| chr1_+_84873913 | 0.14 |
ENST00000370662.3 |
DNASE2B |
deoxyribonuclease II beta |
| chr3_-_48481518 | 0.13 |
ENST00000412398.2 ENST00000395696.1 |
CCDC51 |
coiled-coil domain containing 51 |
| chr12_+_9144626 | 0.13 |
ENST00000543895.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr1_+_100315613 | 0.13 |
ENST00000361915.3 |
AGL |
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr2_-_118943930 | 0.13 |
ENST00000449075.1 ENST00000414886.1 ENST00000449819.1 |
AC093901.1 |
AC093901.1 |
| chr17_+_11924129 | 0.13 |
ENST00000353533.5 ENST00000415385.3 |
MAP2K4 |
mitogen-activated protein kinase kinase 4 |
| chr11_+_60145997 | 0.12 |
ENST00000530614.1 ENST00000530027.1 ENST00000530234.2 ENST00000528215.1 ENST00000531787.1 |
MS4A7 MS4A14 |
membrane-spanning 4-domains, subfamily A, member 7 membrane-spanning 4-domains, subfamily A, member 14 |
| chr2_+_234627424 | 0.12 |
ENST00000373409.3 |
UGT1A4 |
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr11_+_60163918 | 0.12 |
ENST00000526375.1 ENST00000531783.1 ENST00000395001.1 |
MS4A14 |
membrane-spanning 4-domains, subfamily A, member 14 |
| chr19_-_15590306 | 0.12 |
ENST00000292609.4 |
PGLYRP2 |
peptidoglycan recognition protein 2 |
| chr10_-_95209 | 0.12 |
ENST00000332708.5 ENST00000309812.4 |
TUBB8 |
tubulin, beta 8 class VIII |
| chr2_-_89327228 | 0.12 |
ENST00000483158.1 |
IGKV3-11 |
immunoglobulin kappa variable 3-11 |
| chr18_+_20513782 | 0.12 |
ENST00000399722.2 ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8 |
retinoblastoma binding protein 8 |
| chr11_+_60163775 | 0.12 |
ENST00000300187.6 ENST00000395005.2 |
MS4A14 |
membrane-spanning 4-domains, subfamily A, member 14 |
| chr15_-_43212996 | 0.12 |
ENST00000567840.1 |
TTBK2 |
tau tubulin kinase 2 |
| chr13_-_46756351 | 0.12 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr6_-_26216872 | 0.12 |
ENST00000244601.3 |
HIST1H2BG |
histone cluster 1, H2bg |
| chr19_+_34895289 | 0.12 |
ENST00000246535.3 |
PDCD2L |
programmed cell death 2-like |
| chr3_+_111260954 | 0.12 |
ENST00000283285.5 |
CD96 |
CD96 molecule |
| chrX_+_99899180 | 0.12 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr8_+_72587535 | 0.11 |
ENST00000519840.1 ENST00000521131.1 |
RP11-1144P22.1 |
RP11-1144P22.1 |
| chr14_+_22670455 | 0.11 |
ENST00000390460.1 |
TRAV26-2 |
T cell receptor alpha variable 26-2 |
| chr3_+_111260856 | 0.11 |
ENST00000352690.4 |
CD96 |
CD96 molecule |
| chr15_-_43212836 | 0.11 |
ENST00000566931.1 ENST00000564431.1 ENST00000567274.1 |
TTBK2 |
tau tubulin kinase 2 |
| chr1_+_174417095 | 0.11 |
ENST00000367685.2 |
GPR52 |
G protein-coupled receptor 52 |
| chr4_-_147043058 | 0.11 |
ENST00000512063.1 ENST00000507726.1 |
RP11-6L6.3 |
long intergenic non-protein coding RNA 1095 |
| chr4_+_128554081 | 0.11 |
ENST00000335251.6 ENST00000296461.5 |
INTU |
inturned planar cell polarity protein |
| chr14_+_105147464 | 0.11 |
ENST00000540171.2 |
RP11-982M15.6 |
RP11-982M15.6 |
| chr11_-_8959758 | 0.11 |
ENST00000531618.1 |
ASCL3 |
achaete-scute family bHLH transcription factor 3 |
| chr2_+_48844937 | 0.10 |
ENST00000448460.1 ENST00000437125.1 ENST00000430487.2 |
GTF2A1L |
general transcription factor IIA, 1-like |
| chr3_-_52488048 | 0.10 |
ENST00000232975.3 |
TNNC1 |
troponin C type 1 (slow) |
| chr6_-_119256311 | 0.10 |
ENST00000316316.6 |
MCM9 |
minichromosome maintenance complex component 9 |
| chr1_+_63063152 | 0.10 |
ENST00000371129.3 |
ANGPTL3 |
angiopoietin-like 3 |
| chr10_+_76871229 | 0.10 |
ENST00000372690.3 |
SAMD8 |
sterile alpha motif domain containing 8 |
| chr17_-_57784755 | 0.10 |
ENST00000537860.1 ENST00000393038.2 ENST00000409433.2 |
PTRH2 |
peptidyl-tRNA hydrolase 2 |
| chr5_+_54398463 | 0.10 |
ENST00000274306.6 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr3_-_113918254 | 0.10 |
ENST00000460779.1 |
DRD3 |
dopamine receptor D3 |
| chr17_+_27181956 | 0.09 |
ENST00000254928.5 ENST00000580917.2 |
ERAL1 |
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr9_+_139717847 | 0.09 |
ENST00000436380.1 |
RABL6 |
RAB, member RAS oncogene family-like 6 |
| chr9_-_21385396 | 0.09 |
ENST00000380206.2 |
IFNA2 |
interferon, alpha 2 |
| chr6_-_135271260 | 0.09 |
ENST00000265605.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr15_-_33360085 | 0.09 |
ENST00000334528.9 |
FMN1 |
formin 1 |
| chr2_+_234637754 | 0.09 |
ENST00000482026.1 ENST00000609767.1 |
UGT1A3 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_+_151009035 | 0.09 |
ENST00000368931.3 |
BNIPL |
BCL2/adenovirus E1B 19kD interacting protein like |
| chr3_-_45838011 | 0.09 |
ENST00000358525.4 ENST00000413781.1 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr18_-_13915530 | 0.09 |
ENST00000327606.3 |
MC2R |
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr3_-_64009102 | 0.09 |
ENST00000478185.1 ENST00000482510.1 ENST00000497323.1 ENST00000492933.1 ENST00000295901.4 |
PSMD6 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr6_-_135271219 | 0.09 |
ENST00000367847.2 ENST00000367845.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr2_+_48844973 | 0.09 |
ENST00000403751.3 |
GTF2A1L |
general transcription factor IIA, 1-like |
| chrX_-_55187531 | 0.09 |
ENST00000489298.1 ENST00000477847.2 |
FAM104B |
family with sequence similarity 104, member B |
| chr16_-_15187865 | 0.09 |
ENST00000327307.7 |
RRN3 |
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr22_+_44464923 | 0.09 |
ENST00000404989.1 |
PARVB |
parvin, beta |
| chr20_+_1184098 | 0.09 |
ENST00000400633.1 |
C20orf202 |
chromosome 20 open reading frame 202 |
| chrX_-_6453159 | 0.09 |
ENST00000381089.3 ENST00000398729.1 |
VCX3A |
variable charge, X-linked 3A |
| chr15_-_81616446 | 0.09 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr2_-_170430366 | 0.09 |
ENST00000453153.2 ENST00000445210.1 |
FASTKD1 |
FAST kinase domains 1 |
| chr4_-_74864386 | 0.09 |
ENST00000296027.4 |
CXCL5 |
chemokine (C-X-C motif) ligand 5 |
| chr11_-_35440579 | 0.09 |
ENST00000606205.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr8_-_141810634 | 0.09 |
ENST00000521986.1 ENST00000523539.1 ENST00000538769.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr19_-_47735918 | 0.09 |
ENST00000449228.1 ENST00000300880.7 ENST00000341983.4 |
BBC3 |
BCL2 binding component 3 |
| chr1_+_173793641 | 0.08 |
ENST00000361951.4 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
| chr20_-_62199427 | 0.08 |
ENST00000427522.2 |
HELZ2 |
helicase with zinc finger 2, transcriptional coactivator |
| chr11_+_5372738 | 0.08 |
ENST00000380219.1 |
OR51B6 |
olfactory receptor, family 51, subfamily B, member 6 |
| chr11_-_110561721 | 0.08 |
ENST00000357139.3 |
ARHGAP20 |
Rho GTPase activating protein 20 |
| chr11_+_3819049 | 0.08 |
ENST00000396986.2 ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2 |
post-GPI attachment to proteins 2 |
| chr3_+_98451275 | 0.08 |
ENST00000265261.6 ENST00000497008.1 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr17_-_41132410 | 0.08 |
ENST00000409446.3 ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L PTGES3L-AARSD1 |
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr17_-_39324424 | 0.08 |
ENST00000391356.2 |
KRTAP4-3 |
keratin associated protein 4-3 |
| chr3_-_45837959 | 0.08 |
ENST00000353278.4 ENST00000456124.2 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr1_-_160492994 | 0.08 |
ENST00000368055.1 ENST00000368057.3 ENST00000368059.3 |
SLAMF6 |
SLAM family member 6 |
| chr1_+_173793777 | 0.08 |
ENST00000239457.5 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
| chr4_-_186877806 | 0.08 |
ENST00000355634.5 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr2_+_1418154 | 0.08 |
ENST00000423320.1 ENST00000382198.1 |
TPO |
thyroid peroxidase |
| chr11_-_118095718 | 0.08 |
ENST00000526620.1 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
| chr12_-_323689 | 0.08 |
ENST00000428720.1 |
SLC6A12 |
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr14_-_106610852 | 0.08 |
ENST00000390603.2 |
IGHV3-15 |
immunoglobulin heavy variable 3-15 |
| chr14_+_22694606 | 0.08 |
ENST00000390463.3 |
TRAV36DV7 |
T cell receptor alpha variable 36/delta variable 7 |
| chr1_-_58716197 | 0.08 |
ENST00000371234.4 |
DAB1 |
Dab, reelin signal transducer, homolog 1 (Drosophila) |
| chr20_-_35807741 | 0.07 |
ENST00000434295.1 ENST00000441008.2 ENST00000400441.3 ENST00000343811.4 |
MROH8 |
maestro heat-like repeat family member 8 |
| chr1_+_156589051 | 0.07 |
ENST00000255039.1 |
HAPLN2 |
hyaluronan and proteoglycan link protein 2 |
| chr17_-_3182268 | 0.07 |
ENST00000408891.2 |
OR3A2 |
olfactory receptor, family 3, subfamily A, member 2 |
| chr17_+_36452989 | 0.07 |
ENST00000312513.5 ENST00000582535.1 |
MRPL45 |
mitochondrial ribosomal protein L45 |
| chr2_+_234526272 | 0.07 |
ENST00000373450.4 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr7_-_102257139 | 0.07 |
ENST00000521076.1 ENST00000462172.1 ENST00000522801.1 ENST00000449970.2 ENST00000262940.7 |
RASA4 |
RAS p21 protein activator 4 |
| chr17_+_68100989 | 0.07 |
ENST00000585558.1 ENST00000392670.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr6_-_31239846 | 0.07 |
ENST00000415537.1 ENST00000376228.5 ENST00000383329.3 |
HLA-C |
major histocompatibility complex, class I, C |
| chr18_+_9885760 | 0.07 |
ENST00000536353.2 ENST00000584255.1 |
TXNDC2 |
thioredoxin domain containing 2 (spermatozoa) |
| chr6_+_84569359 | 0.07 |
ENST00000369681.5 ENST00000369679.4 |
CYB5R4 |
cytochrome b5 reductase 4 |
| chr6_+_26217159 | 0.07 |
ENST00000303910.2 |
HIST1H2AE |
histone cluster 1, H2ae |
| chr1_+_87012753 | 0.07 |
ENST00000370563.3 |
CLCA4 |
chloride channel accessory 4 |
| chr9_-_71155783 | 0.07 |
ENST00000377311.3 |
TMEM252 |
transmembrane protein 252 |
| chr12_+_25205446 | 0.07 |
ENST00000557489.1 ENST00000354454.3 ENST00000536173.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr11_-_116708302 | 0.07 |
ENST00000375320.1 ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1 |
apolipoprotein A-I |
| chr18_+_616672 | 0.07 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr12_+_70219052 | 0.07 |
ENST00000552032.2 ENST00000547771.2 |
MYRFL |
myelin regulatory factor-like |
| chr16_-_20709066 | 0.07 |
ENST00000520010.1 |
ACSM1 |
acyl-CoA synthetase medium-chain family member 1 |
| chr5_-_10308125 | 0.07 |
ENST00000296658.3 |
CMBL |
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr10_+_135340859 | 0.07 |
ENST00000252945.3 ENST00000421586.1 ENST00000418356.1 |
CYP2E1 |
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr19_+_10765003 | 0.07 |
ENST00000407004.3 ENST00000589998.1 ENST00000589600.1 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
| chr2_+_102927962 | 0.07 |
ENST00000233954.1 ENST00000393393.3 ENST00000410040.1 |
IL1RL1 IL18R1 |
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr3_-_123710893 | 0.07 |
ENST00000467907.1 ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1 |
rhophilin associated tail protein 1 |
| chr14_-_107013465 | 0.06 |
ENST00000390625.2 |
IGHV3-49 |
immunoglobulin heavy variable 3-49 |
| chr15_-_76304731 | 0.06 |
ENST00000394907.3 |
NRG4 |
neuregulin 4 |
| chr18_-_19748379 | 0.06 |
ENST00000579431.1 |
GATA6-AS1 |
GATA6 antisense RNA 1 (head to head) |
| chr5_-_1345199 | 0.06 |
ENST00000320895.5 |
CLPTM1L |
CLPTM1-like |
| chrX_-_118699325 | 0.06 |
ENST00000320339.4 ENST00000371594.4 ENST00000536133.1 |
CXorf56 |
chromosome X open reading frame 56 |
| chr2_+_113816215 | 0.06 |
ENST00000346807.3 |
IL36RN |
interleukin 36 receptor antagonist |
| chr19_+_3933579 | 0.06 |
ENST00000593949.1 |
NMRK2 |
nicotinamide riboside kinase 2 |
| chr18_-_19748331 | 0.06 |
ENST00000584201.1 |
GATA6-AS1 |
GATA6 antisense RNA 1 (head to head) |
| chr2_+_102413726 | 0.06 |
ENST00000350878.4 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr17_-_36997708 | 0.06 |
ENST00000398575.4 |
C17orf98 |
chromosome 17 open reading frame 98 |
| chr19_-_16606988 | 0.06 |
ENST00000269881.3 |
CALR3 |
calreticulin 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.6 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 | 1.0 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.5 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 4.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.3 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.2 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.1 | GO:0010390 | histone monoubiquitination(GO:0010390) histone H2A ubiquitination(GO:0033522) histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 0.3 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.3 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.1 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.1 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.3 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.7 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 1.4 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.0 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.0 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.2 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:1902714 | antifungal humoral response(GO:0019732) negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.6 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.1 | 0.7 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 1.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.1 | 0.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.6 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein antigen binding(GO:1990405) |
| 0.1 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.3 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 3.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 1.4 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.0 | 0.1 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0033677 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |