Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
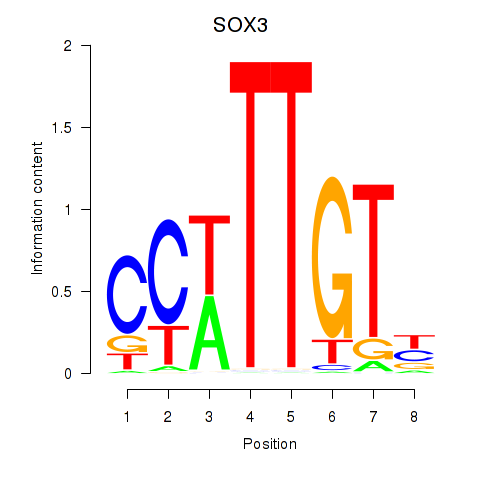
Results for SOX3_SOX2
Z-value: 0.05
Transcription factors associated with SOX3_SOX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX3
|
ENSG00000134595.6 | SOX3 |
|
SOX2
|
ENSG00000181449.2 | SOX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX3 | hg19_v2_chrX_-_139587225_139587234 | -0.88 | 3.9e-03 | Click! |
| SOX2 | hg19_v2_chr3_+_181429704_181429722 | 0.69 | 6.0e-02 | Click! |
Activity profile of SOX3_SOX2 motif
Sorted Z-values of SOX3_SOX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX3_SOX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_138453606 | 1.15 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr4_-_138453559 | 0.98 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr6_+_72596604 | 0.80 |
ENST00000348717.5 ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr8_-_124553437 | 0.73 |
ENST00000517956.1 ENST00000443022.2 |
FBXO32 |
F-box protein 32 |
| chr4_-_186877502 | 0.65 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr22_+_27053422 | 0.62 |
ENST00000413665.1 ENST00000421151.1 ENST00000456129.1 ENST00000430080.1 |
MIAT |
myocardial infarction associated transcript (non-protein coding) |
| chr8_-_22550815 | 0.58 |
ENST00000317216.2 |
EGR3 |
early growth response 3 |
| chr16_-_4987065 | 0.57 |
ENST00000590782.2 ENST00000345988.2 |
PPL |
periplakin |
| chr4_-_186732048 | 0.56 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr9_-_131940526 | 0.53 |
ENST00000372491.2 |
IER5L |
immediate early response 5-like |
| chr2_+_210444142 | 0.51 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr11_-_94965667 | 0.51 |
ENST00000542176.1 ENST00000278499.2 |
SESN3 |
sestrin 3 |
| chr3_-_114343768 | 0.48 |
ENST00000393785.2 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr9_+_101705893 | 0.48 |
ENST00000375001.3 |
COL15A1 |
collagen, type XV, alpha 1 |
| chr8_+_104384616 | 0.47 |
ENST00000520337.1 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr6_+_30848557 | 0.46 |
ENST00000460944.2 ENST00000324771.8 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr1_+_215256467 | 0.44 |
ENST00000391894.2 ENST00000444842.2 |
KCNK2 |
potassium channel, subfamily K, member 2 |
| chr3_-_18466026 | 0.44 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr22_+_27053190 | 0.42 |
ENST00000439738.1 ENST00000422403.1 ENST00000436238.1 ENST00000425476.1 ENST00000455640.1 ENST00000451141.1 ENST00000452429.1 ENST00000423278.1 |
MIAT |
myocardial infarction associated transcript (non-protein coding) |
| chr9_-_123476612 | 0.42 |
ENST00000426959.1 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr9_-_123476719 | 0.41 |
ENST00000373930.3 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr4_+_62067860 | 0.41 |
ENST00000514591.1 |
LPHN3 |
latrophilin 3 |
| chr20_-_39317868 | 0.40 |
ENST00000373313.2 |
MAFB |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr19_+_45409011 | 0.39 |
ENST00000252486.4 ENST00000446996.1 ENST00000434152.1 |
APOE |
apolipoprotein E |
| chr15_-_93632421 | 0.39 |
ENST00000329082.7 |
RGMA |
repulsive guidance molecule family member a |
| chrX_+_55744228 | 0.38 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr1_+_145727681 | 0.38 |
ENST00000417171.1 ENST00000451928.2 |
PDZK1 |
PDZ domain containing 1 |
| chr7_+_16793160 | 0.38 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr4_+_95679072 | 0.38 |
ENST00000515059.1 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chrX_+_55744166 | 0.38 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr4_-_159080806 | 0.37 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr11_-_111794446 | 0.36 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr3_-_48470838 | 0.36 |
ENST00000358459.4 ENST00000358536.4 |
PLXNB1 |
plexin B1 |
| chr1_-_32801825 | 0.36 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr4_+_41614720 | 0.36 |
ENST00000509277.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr9_-_95298314 | 0.35 |
ENST00000344604.5 ENST00000375540.1 |
ECM2 |
extracellular matrix protein 2, female organ and adipocyte specific |
| chr14_-_22005018 | 0.35 |
ENST00000546363.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr11_-_111781554 | 0.35 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr1_-_156828810 | 0.35 |
ENST00000368195.3 |
INSRR |
insulin receptor-related receptor |
| chr6_+_72596406 | 0.34 |
ENST00000491071.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr10_-_70092671 | 0.34 |
ENST00000358769.2 ENST00000432941.1 ENST00000495025.2 |
PBLD |
phenazine biosynthesis-like protein domain containing |
| chr8_-_17579726 | 0.33 |
ENST00000381861.3 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr12_-_7281469 | 0.33 |
ENST00000542370.1 ENST00000266560.3 |
RBP5 |
retinol binding protein 5, cellular |
| chr10_+_60759378 | 0.33 |
ENST00000432535.1 |
LINC00844 |
long intergenic non-protein coding RNA 844 |
| chr1_+_200993071 | 0.33 |
ENST00000446333.1 ENST00000458003.1 |
RP11-168O16.1 |
RP11-168O16.1 |
| chr1_+_33722080 | 0.33 |
ENST00000483388.1 ENST00000539719.1 |
ZNF362 |
zinc finger protein 362 |
| chr15_-_52944231 | 0.33 |
ENST00000546305.2 |
FAM214A |
family with sequence similarity 214, member A |
| chr1_+_11751748 | 0.32 |
ENST00000294485.5 |
DRAXIN |
dorsal inhibitory axon guidance protein |
| chr4_-_2264015 | 0.32 |
ENST00000337190.2 |
MXD4 |
MAX dimerization protein 4 |
| chr5_-_175964366 | 0.32 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr11_-_111781610 | 0.32 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr3_-_58563094 | 0.31 |
ENST00000464064.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr11_-_111781454 | 0.31 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr15_-_82338460 | 0.31 |
ENST00000558133.1 ENST00000329713.4 |
MEX3B |
mex-3 RNA binding family member B |
| chr15_+_22382382 | 0.31 |
ENST00000328795.4 |
OR4N4 |
olfactory receptor, family 4, subfamily N, member 4 |
| chrX_+_84499081 | 0.30 |
ENST00000276123.3 |
ZNF711 |
zinc finger protein 711 |
| chr16_-_11730213 | 0.30 |
ENST00000576334.1 ENST00000574848.1 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr1_+_197886461 | 0.29 |
ENST00000367388.3 ENST00000337020.2 ENST00000367387.4 |
LHX9 |
LIM homeobox 9 |
| chr8_-_21988558 | 0.29 |
ENST00000312841.8 |
HR |
hair growth associated |
| chr7_-_19813192 | 0.29 |
ENST00000422233.1 ENST00000433641.1 |
TMEM196 |
transmembrane protein 196 |
| chr19_-_20844343 | 0.29 |
ENST00000595405.1 |
ZNF626 |
zinc finger protein 626 |
| chr11_+_57365150 | 0.29 |
ENST00000457869.1 ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1 |
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr7_+_20370746 | 0.29 |
ENST00000222573.4 |
ITGB8 |
integrin, beta 8 |
| chr2_-_1748214 | 0.29 |
ENST00000433670.1 ENST00000425171.1 ENST00000252804.4 |
PXDN |
peroxidasin homolog (Drosophila) |
| chr3_-_87040233 | 0.28 |
ENST00000398399.2 |
VGLL3 |
vestigial like 3 (Drosophila) |
| chr2_-_175869936 | 0.28 |
ENST00000409900.3 |
CHN1 |
chimerin 1 |
| chr2_+_210517895 | 0.28 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr9_+_139874683 | 0.27 |
ENST00000444903.1 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chr6_+_112375275 | 0.27 |
ENST00000368666.2 ENST00000604763.1 ENST00000230529.5 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
| chr17_-_9940058 | 0.27 |
ENST00000585266.1 |
GAS7 |
growth arrest-specific 7 |
| chrX_-_142722897 | 0.27 |
ENST00000338017.4 |
SLITRK4 |
SLIT and NTRK-like family, member 4 |
| chr4_-_175443943 | 0.27 |
ENST00000296522.6 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr17_+_7608511 | 0.27 |
ENST00000226091.2 |
EFNB3 |
ephrin-B3 |
| chr3_+_39509070 | 0.26 |
ENST00000354668.4 ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP |
myelin-associated oligodendrocyte basic protein |
| chr20_-_50418947 | 0.26 |
ENST00000371539.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr21_+_27011584 | 0.26 |
ENST00000400532.1 ENST00000480456.1 ENST00000312957.5 |
JAM2 |
junctional adhesion molecule 2 |
| chr17_+_1674982 | 0.25 |
ENST00000572048.1 ENST00000573763.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr20_-_50418972 | 0.25 |
ENST00000395997.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr20_-_50419055 | 0.25 |
ENST00000217086.4 |
SALL4 |
spalt-like transcription factor 4 |
| chrX_+_80457442 | 0.25 |
ENST00000373212.5 |
SH3BGRL |
SH3 domain binding glutamic acid-rich protein like |
| chr12_+_31079652 | 0.25 |
ENST00000546076.1 ENST00000535215.1 ENST00000544427.1 ENST00000261177.9 |
TSPAN11 |
tetraspanin 11 |
| chr1_-_156217829 | 0.25 |
ENST00000356983.2 ENST00000335852.1 ENST00000340183.5 ENST00000540423.1 |
PAQR6 |
progestin and adipoQ receptor family member VI |
| chr1_+_164529004 | 0.25 |
ENST00000559240.1 ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr19_-_55691377 | 0.25 |
ENST00000589172.1 |
SYT5 |
synaptotagmin V |
| chr14_+_96505659 | 0.25 |
ENST00000555004.1 |
C14orf132 |
chromosome 14 open reading frame 132 |
| chr6_-_128841503 | 0.25 |
ENST00000368215.3 ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
| chr1_-_156217822 | 0.24 |
ENST00000368270.1 |
PAQR6 |
progestin and adipoQ receptor family member VI |
| chr4_+_86699834 | 0.24 |
ENST00000395183.2 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr8_-_108510224 | 0.24 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr12_+_80750134 | 0.24 |
ENST00000546620.1 |
OTOGL |
otogelin-like |
| chr5_+_140593509 | 0.24 |
ENST00000341948.4 |
PCDHB13 |
protocadherin beta 13 |
| chr4_-_90757364 | 0.24 |
ENST00000508895.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_-_3528034 | 0.24 |
ENST00000356575.4 |
MEGF6 |
multiple EGF-like-domains 6 |
| chr14_-_91884115 | 0.24 |
ENST00000389857.6 |
CCDC88C |
coiled-coil domain containing 88C |
| chr1_-_161015752 | 0.23 |
ENST00000435396.1 ENST00000368021.3 |
USF1 |
upstream transcription factor 1 |
| chr4_-_5890145 | 0.23 |
ENST00000397890.2 |
CRMP1 |
collapsin response mediator protein 1 |
| chr3_+_149689066 | 0.23 |
ENST00000593416.1 |
AC117395.1 |
LOC646903 protein; Uncharacterized protein |
| chr1_-_156217875 | 0.23 |
ENST00000292291.5 |
PAQR6 |
progestin and adipoQ receptor family member VI |
| chr2_+_105050794 | 0.23 |
ENST00000429464.1 ENST00000414442.1 ENST00000447380.1 |
AC013402.2 |
long intergenic non-protein coding RNA 1102 |
| chr2_+_210444748 | 0.23 |
ENST00000392194.1 |
MAP2 |
microtubule-associated protein 2 |
| chr16_-_31085033 | 0.23 |
ENST00000414399.1 |
ZNF668 |
zinc finger protein 668 |
| chr5_+_15500280 | 0.23 |
ENST00000504595.1 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
| chr10_-_70092635 | 0.23 |
ENST00000309049.4 |
PBLD |
phenazine biosynthesis-like protein domain containing |
| chr5_-_39425290 | 0.22 |
ENST00000545653.1 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr18_+_6729698 | 0.22 |
ENST00000383472.4 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr11_-_117747434 | 0.22 |
ENST00000529335.2 ENST00000530956.1 ENST00000260282.4 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr6_-_46459099 | 0.22 |
ENST00000371374.1 |
RCAN2 |
regulator of calcineurin 2 |
| chr8_-_48651648 | 0.22 |
ENST00000408965.3 |
CEBPD |
CCAAT/enhancer binding protein (C/EBP), delta |
| chr5_+_140235469 | 0.22 |
ENST00000506939.2 ENST00000307360.5 |
PCDHA10 |
protocadherin alpha 10 |
| chr5_+_148960931 | 0.22 |
ENST00000333677.6 |
ARHGEF37 |
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr3_-_114790179 | 0.22 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr12_+_94542459 | 0.22 |
ENST00000258526.4 |
PLXNC1 |
plexin C1 |
| chr2_+_30454390 | 0.22 |
ENST00000395323.3 ENST00000406087.1 ENST00000404397.1 |
LBH |
limb bud and heart development |
| chr6_-_24877490 | 0.22 |
ENST00000540914.1 ENST00000378023.4 |
FAM65B |
family with sequence similarity 65, member B |
| chr1_-_46598371 | 0.22 |
ENST00000372006.1 ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr16_-_28550348 | 0.22 |
ENST00000324873.6 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
| chr7_+_141463897 | 0.22 |
ENST00000247879.2 |
TAS2R3 |
taste receptor, type 2, member 3 |
| chr6_+_126240442 | 0.22 |
ENST00000448104.1 ENST00000438495.1 ENST00000444128.1 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr5_-_39425222 | 0.21 |
ENST00000320816.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr3_+_44596679 | 0.21 |
ENST00000426540.1 ENST00000431636.1 ENST00000341840.3 ENST00000273320.3 |
ZKSCAN7 |
zinc finger with KRAB and SCAN domains 7 |
| chr14_+_91580777 | 0.21 |
ENST00000525393.2 ENST00000428926.2 ENST00000517362.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chrX_+_100333709 | 0.21 |
ENST00000372930.4 |
TMEM35 |
transmembrane protein 35 |
| chr19_-_7553852 | 0.21 |
ENST00000593547.1 |
PEX11G |
peroxisomal biogenesis factor 11 gamma |
| chr10_+_72972281 | 0.21 |
ENST00000335350.6 |
UNC5B |
unc-5 homolog B (C. elegans) |
| chr14_+_91580708 | 0.21 |
ENST00000518868.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr1_-_3447967 | 0.21 |
ENST00000294599.4 |
MEGF6 |
multiple EGF-like-domains 6 |
| chr1_+_144339738 | 0.21 |
ENST00000538264.1 |
AL592284.1 |
Protein LOC642441 |
| chr1_-_1243392 | 0.20 |
ENST00000354700.5 |
ACAP3 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr4_-_90756769 | 0.20 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_-_175443788 | 0.20 |
ENST00000541923.1 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr14_-_73493784 | 0.20 |
ENST00000553891.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr3_+_133118839 | 0.20 |
ENST00000302334.2 |
BFSP2 |
beaded filament structural protein 2, phakinin |
| chr22_-_32555275 | 0.20 |
ENST00000382097.3 |
C22orf42 |
chromosome 22 open reading frame 42 |
| chr14_+_91581011 | 0.20 |
ENST00000523894.1 ENST00000522322.1 ENST00000523771.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr7_+_70597109 | 0.19 |
ENST00000333538.5 |
WBSCR17 |
Williams-Beuren syndrome chromosome region 17 |
| chr12_+_31477250 | 0.19 |
ENST00000313737.4 |
AC024940.1 |
AC024940.1 |
| chr14_+_95078714 | 0.19 |
ENST00000393078.3 ENST00000393080.4 ENST00000467132.1 |
SERPINA3 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr12_-_10588539 | 0.19 |
ENST00000381902.2 ENST00000381901.1 ENST00000539033.1 |
KLRC2 NKG2-E |
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chr11_-_10829851 | 0.19 |
ENST00000532082.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr1_-_153283194 | 0.19 |
ENST00000290722.1 |
PGLYRP3 |
peptidoglycan recognition protein 3 |
| chrX_+_102469997 | 0.18 |
ENST00000372695.5 ENST00000372691.3 |
BEX4 |
brain expressed, X-linked 4 |
| chr6_+_89790490 | 0.18 |
ENST00000336032.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr3_+_183353356 | 0.18 |
ENST00000242810.6 ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24 |
kelch-like family member 24 |
| chr8_-_93115445 | 0.18 |
ENST00000523629.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr6_+_89790459 | 0.18 |
ENST00000369472.1 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr11_+_1889880 | 0.18 |
ENST00000405957.2 |
LSP1 |
lymphocyte-specific protein 1 |
| chr2_+_74685527 | 0.18 |
ENST00000393972.3 ENST00000409737.1 ENST00000428943.1 |
WBP1 |
WW domain binding protein 1 |
| chrX_+_69488174 | 0.18 |
ENST00000480877.2 ENST00000307959.8 |
ARR3 |
arrestin 3, retinal (X-arrestin) |
| chr3_-_48700310 | 0.18 |
ENST00000164024.4 ENST00000544264.1 |
CELSR3 |
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr5_-_39425068 | 0.18 |
ENST00000515700.1 ENST00000339788.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr2_+_181845298 | 0.18 |
ENST00000410062.4 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
| chr13_+_31506818 | 0.18 |
ENST00000380473.3 |
TEX26 |
testis expressed 26 |
| chr4_-_175443484 | 0.18 |
ENST00000514584.1 ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr4_+_15480828 | 0.18 |
ENST00000389652.5 |
CC2D2A |
coiled-coil and C2 domain containing 2A |
| chr14_-_73493825 | 0.18 |
ENST00000318876.5 ENST00000556143.1 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
| chr15_+_34310428 | 0.18 |
ENST00000557872.1 |
CHRM5 |
cholinergic receptor, muscarinic 5 |
| chr4_+_41258786 | 0.18 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chrX_-_13956497 | 0.18 |
ENST00000398361.3 |
GPM6B |
glycoprotein M6B |
| chr8_-_120685608 | 0.17 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chrX_+_150866779 | 0.17 |
ENST00000370353.3 |
PRRG3 |
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr3_+_39509163 | 0.17 |
ENST00000436143.2 ENST00000441980.2 ENST00000311042.6 |
MOBP |
myelin-associated oligodendrocyte basic protein |
| chr2_-_238499303 | 0.17 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chrX_+_103031421 | 0.17 |
ENST00000433491.1 ENST00000418604.1 ENST00000443502.1 |
PLP1 |
proteolipid protein 1 |
| chr2_+_177015950 | 0.17 |
ENST00000306324.3 |
HOXD4 |
homeobox D4 |
| chr11_-_2162468 | 0.17 |
ENST00000434045.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr1_+_244998602 | 0.17 |
ENST00000411948.2 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
| chr3_-_116163830 | 0.17 |
ENST00000333617.4 |
LSAMP |
limbic system-associated membrane protein |
| chr10_+_70320413 | 0.17 |
ENST00000373644.4 |
TET1 |
tet methylcytosine dioxygenase 1 |
| chr12_-_95611149 | 0.17 |
ENST00000549499.1 ENST00000343958.4 ENST00000546711.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr1_-_161014731 | 0.17 |
ENST00000368020.1 |
USF1 |
upstream transcription factor 1 |
| chr2_-_166060571 | 0.17 |
ENST00000360093.3 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr9_-_130712995 | 0.17 |
ENST00000373084.4 |
FAM102A |
family with sequence similarity 102, member A |
| chrX_+_103031758 | 0.17 |
ENST00000303958.2 ENST00000361621.2 |
PLP1 |
proteolipid protein 1 |
| chr22_+_30792846 | 0.17 |
ENST00000312932.9 ENST00000428195.1 |
SEC14L2 |
SEC14-like 2 (S. cerevisiae) |
| chr9_+_2622085 | 0.17 |
ENST00000382099.2 |
VLDLR |
very low density lipoprotein receptor |
| chr13_-_28674693 | 0.16 |
ENST00000537084.1 ENST00000241453.7 ENST00000380982.4 |
FLT3 |
fms-related tyrosine kinase 3 |
| chr17_+_4046462 | 0.16 |
ENST00000577075.2 ENST00000575251.1 ENST00000301391.3 |
CYB5D2 |
cytochrome b5 domain containing 2 |
| chr12_+_133707204 | 0.16 |
ENST00000426665.2 |
ZNF10 |
zinc finger protein 10 |
| chr1_+_164528866 | 0.16 |
ENST00000420696.2 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr12_-_52887034 | 0.16 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr1_-_108231101 | 0.16 |
ENST00000544443.1 ENST00000415432.2 |
VAV3 |
vav 3 guanine nucleotide exchange factor |
| chr16_+_57673430 | 0.16 |
ENST00000540164.2 ENST00000568531.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr2_+_223536428 | 0.16 |
ENST00000446656.3 |
MOGAT1 |
monoacylglycerol O-acyltransferase 1 |
| chr12_+_95611569 | 0.16 |
ENST00000261219.6 ENST00000551472.1 ENST00000552821.1 |
VEZT |
vezatin, adherens junctions transmembrane protein |
| chr16_-_74455290 | 0.16 |
ENST00000339953.5 |
CLEC18B |
C-type lectin domain family 18, member B |
| chr2_-_166060552 | 0.16 |
ENST00000283254.7 ENST00000453007.1 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr20_+_30598231 | 0.16 |
ENST00000300415.8 ENST00000262659.8 |
CCM2L |
cerebral cavernous malformation 2-like |
| chr10_-_49482907 | 0.16 |
ENST00000374201.3 ENST00000407470.4 |
FRMPD2 |
FERM and PDZ domain containing 2 |
| chr7_-_99869799 | 0.16 |
ENST00000436886.2 |
GATS |
GATS, stromal antigen 3 opposite strand |
| chr3_-_11623804 | 0.16 |
ENST00000451674.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr22_-_32341336 | 0.16 |
ENST00000248984.3 |
C22orf24 |
chromosome 22 open reading frame 24 |
| chr16_+_54964740 | 0.16 |
ENST00000394636.4 |
IRX5 |
iroquois homeobox 5 |
| chr1_-_1334685 | 0.16 |
ENST00000400809.3 ENST00000408918.4 |
CCNL2 |
cyclin L2 |
| chr17_+_27920486 | 0.15 |
ENST00000394859.3 |
ANKRD13B |
ankyrin repeat domain 13B |
| chr19_+_1491144 | 0.15 |
ENST00000233596.3 |
REEP6 |
receptor accessory protein 6 |
| chr1_-_17307173 | 0.15 |
ENST00000438542.1 ENST00000375535.3 |
MFAP2 |
microfibrillar-associated protein 2 |
| chr1_+_235491714 | 0.15 |
ENST00000471812.1 ENST00000358966.2 ENST00000282841.5 ENST00000391855.2 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
| chr2_+_111880242 | 0.15 |
ENST00000393252.3 |
BCL2L11 |
BCL2-like 11 (apoptosis facilitator) |
| chrX_-_139866723 | 0.15 |
ENST00000370532.2 |
CDR1 |
cerebellar degeneration-related protein 1, 34kDa |
| chr20_+_61448376 | 0.15 |
ENST00000343916.3 |
COL9A3 |
collagen, type IX, alpha 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 0.6 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 0.6 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.2 | 0.6 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.4 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.4 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.1 | 0.4 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.4 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.3 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.1 | 0.8 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.4 | GO:1902731 | negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.2 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.1 | 0.2 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 1.4 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 1.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.6 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 0.6 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 0.5 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.1 | 0.4 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.7 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.1 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.2 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 0.2 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.1 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.1 | GO:0035801 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.1 | 0.2 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.3 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.2 | GO:1904300 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.2 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.3 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0072019 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.1 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.3 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.0 | GO:0051354 | negative regulation of oxidoreductase activity(GO:0051354) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.2 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 1.1 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.1 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:0048525 | negative regulation of viral process(GO:0048525) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.2 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.2 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:2000979 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.0 | 0.0 | GO:0070662 | mast cell proliferation(GO:0070662) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.0 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.2 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 3.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0061183 | Spemann organizer formation(GO:0060061) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.1 | GO:1903788 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.3 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.1 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.1 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0039506 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.0 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 | 0.0 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.4 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.3 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.0 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.0 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.0 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.0 | GO:0060458 | right lung development(GO:0060458) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.2 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.0 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.0 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 1.0 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.0 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.0 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.0 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.0 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.1 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.1 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.0 | 0.5 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 0.5 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.8 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 1.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.6 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.5 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.0 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.0 | GO:0019031 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.7 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.4 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.1 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.3 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.4 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.2 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.3 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.2 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.1 | 0.2 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 1.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.0 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.0 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.0 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |