Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011): : averaged replicates
Navigation
Downloads
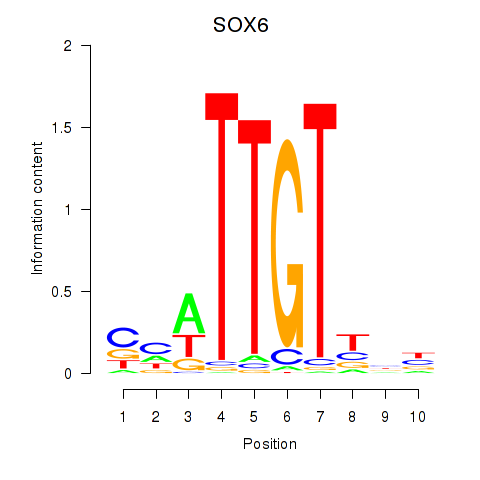
Results for SOX6
Z-value: 0.28
Transcription factors associated with SOX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX6
|
ENSG00000110693.11 | SOX6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX6 | hg19_v2_chr11_-_16430399_16430440 | -0.09 | 8.4e-01 | Click! |
Activity profile of SOX6 motif
Sorted Z-values of SOX6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_70916119 | 0.26 |
ENST00000246896.3 ENST00000511674.1 |
HTN1 |
histatin 1 |
| chr6_+_72596604 | 0.20 |
ENST00000348717.5 ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr8_-_120685608 | 0.18 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr11_+_19798964 | 0.15 |
ENST00000527559.2 |
NAV2 |
neuron navigator 2 |
| chr10_-_25241499 | 0.15 |
ENST00000376378.1 ENST00000376376.3 ENST00000320152.6 |
PRTFDC1 |
phosphoribosyl transferase domain containing 1 |
| chr17_-_19648683 | 0.14 |
ENST00000573368.1 ENST00000457500.2 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr11_+_19799327 | 0.13 |
ENST00000540292.1 |
NAV2 |
neuron navigator 2 |
| chr4_-_138453606 | 0.13 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr9_+_101705893 | 0.13 |
ENST00000375001.3 |
COL15A1 |
collagen, type XV, alpha 1 |
| chr17_-_19648916 | 0.12 |
ENST00000444455.1 ENST00000439102.2 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr5_-_157002749 | 0.12 |
ENST00000517905.1 ENST00000430702.2 ENST00000394020.1 |
ADAM19 |
ADAM metallopeptidase domain 19 |
| chr5_-_157002775 | 0.11 |
ENST00000257527.4 |
ADAM19 |
ADAM metallopeptidase domain 19 |
| chr13_-_24007815 | 0.11 |
ENST00000382298.3 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr20_+_61448376 | 0.11 |
ENST00000343916.3 |
COL9A3 |
collagen, type IX, alpha 3 |
| chr2_-_37899323 | 0.10 |
ENST00000295324.3 ENST00000457889.1 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
| chr16_+_15528332 | 0.10 |
ENST00000566490.1 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr19_-_17375541 | 0.10 |
ENST00000252597.3 |
USHBP1 |
Usher syndrome 1C binding protein 1 |
| chr5_-_39425290 | 0.10 |
ENST00000545653.1 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr15_+_89631381 | 0.10 |
ENST00000352732.5 |
ABHD2 |
abhydrolase domain containing 2 |
| chr5_-_39425222 | 0.10 |
ENST00000320816.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr5_+_140749803 | 0.10 |
ENST00000576222.1 |
PCDHGB3 |
protocadherin gamma subfamily B, 3 |
| chr12_-_45270151 | 0.09 |
ENST00000429094.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr12_-_45270077 | 0.09 |
ENST00000551601.1 ENST00000549027.1 ENST00000452445.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr9_+_139874683 | 0.09 |
ENST00000444903.1 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chr5_-_39425068 | 0.09 |
ENST00000515700.1 ENST00000339788.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr15_+_89631647 | 0.09 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr2_-_86564776 | 0.09 |
ENST00000165698.5 ENST00000541910.1 ENST00000535845.1 |
REEP1 |
receptor accessory protein 1 |
| chr3_-_114790179 | 0.09 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr16_-_73082274 | 0.09 |
ENST00000268489.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr12_+_32260085 | 0.09 |
ENST00000548411.1 ENST00000281474.5 ENST00000551086.1 |
BICD1 |
bicaudal D homolog 1 (Drosophila) |
| chr1_-_156828810 | 0.09 |
ENST00000368195.3 |
INSRR |
insulin receptor-related receptor |
| chr19_-_5340730 | 0.09 |
ENST00000372412.4 ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
| chr4_+_55524085 | 0.09 |
ENST00000412167.2 ENST00000288135.5 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr12_-_10151773 | 0.08 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr3_+_133118839 | 0.08 |
ENST00000302334.2 |
BFSP2 |
beaded filament structural protein 2, phakinin |
| chr3_-_167813132 | 0.08 |
ENST00000309027.4 |
GOLIM4 |
golgi integral membrane protein 4 |
| chr2_-_166060571 | 0.08 |
ENST00000360093.3 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr15_+_85923856 | 0.07 |
ENST00000560302.1 ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr2_-_166060552 | 0.07 |
ENST00000283254.7 ENST00000453007.1 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr15_+_85923797 | 0.07 |
ENST00000559362.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr6_-_24877490 | 0.07 |
ENST00000540914.1 ENST00000378023.4 |
FAM65B |
family with sequence similarity 65, member B |
| chr11_-_82708519 | 0.07 |
ENST00000534301.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr11_+_46402482 | 0.07 |
ENST00000441869.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr1_+_93645314 | 0.07 |
ENST00000343253.7 |
CCDC18 |
coiled-coil domain containing 18 |
| chr14_+_100070869 | 0.07 |
ENST00000502101.2 |
RP11-543C4.1 |
RP11-543C4.1 |
| chr20_-_3996036 | 0.07 |
ENST00000336095.6 |
RNF24 |
ring finger protein 24 |
| chr5_+_140792614 | 0.07 |
ENST00000398610.2 |
PCDHGA10 |
protocadherin gamma subfamily A, 10 |
| chr16_+_23765948 | 0.07 |
ENST00000300113.2 |
CHP2 |
calcineurin-like EF-hand protein 2 |
| chr4_-_138453559 | 0.07 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr11_-_82708435 | 0.06 |
ENST00000525117.1 ENST00000532548.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr10_+_70480963 | 0.06 |
ENST00000265872.6 ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1 |
cell division cycle and apoptosis regulator 1 |
| chr5_-_124081008 | 0.06 |
ENST00000306315.5 |
ZNF608 |
zinc finger protein 608 |
| chr1_-_93645818 | 0.06 |
ENST00000370280.1 ENST00000479918.1 |
TMED5 |
transmembrane emp24 protein transport domain containing 5 |
| chr18_+_7946941 | 0.06 |
ENST00000444013.1 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
| chr16_+_3704822 | 0.06 |
ENST00000414110.2 |
DNASE1 |
deoxyribonuclease I |
| chr11_+_196738 | 0.06 |
ENST00000325113.4 ENST00000342593.5 |
ODF3 |
outer dense fiber of sperm tails 3 |
| chr1_+_197886461 | 0.06 |
ENST00000367388.3 ENST00000337020.2 ENST00000367387.4 |
LHX9 |
LIM homeobox 9 |
| chr18_+_6729698 | 0.06 |
ENST00000383472.4 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr2_+_61293021 | 0.06 |
ENST00000402291.1 |
KIAA1841 |
KIAA1841 |
| chr1_+_228337553 | 0.06 |
ENST00000366714.2 |
GJC2 |
gap junction protein, gamma 2, 47kDa |
| chr19_-_51472823 | 0.06 |
ENST00000310157.2 |
KLK6 |
kallikrein-related peptidase 6 |
| chr1_-_201476274 | 0.06 |
ENST00000340006.2 |
CSRP1 |
cysteine and glycine-rich protein 1 |
| chr6_+_114178512 | 0.05 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr4_+_26585538 | 0.05 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr13_-_110959478 | 0.05 |
ENST00000543140.1 ENST00000375820.4 |
COL4A1 |
collagen, type IV, alpha 1 |
| chr19_-_17375527 | 0.05 |
ENST00000431146.2 ENST00000594190.1 |
USHBP1 |
Usher syndrome 1C binding protein 1 |
| chr1_+_150039369 | 0.05 |
ENST00000369130.3 ENST00000369128.5 |
VPS45 |
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr6_-_109777128 | 0.05 |
ENST00000358807.3 ENST00000358577.3 |
MICAL1 |
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr15_-_82338460 | 0.05 |
ENST00000558133.1 ENST00000329713.4 |
MEX3B |
mex-3 RNA binding family member B |
| chr4_-_146101304 | 0.05 |
ENST00000447906.2 |
OTUD4 |
OTU domain containing 4 |
| chr17_-_9940058 | 0.05 |
ENST00000585266.1 |
GAS7 |
growth arrest-specific 7 |
| chr14_-_106518922 | 0.05 |
ENST00000390598.2 |
IGHV3-7 |
immunoglobulin heavy variable 3-7 |
| chr17_-_14683517 | 0.05 |
ENST00000379640.1 |
AC005863.1 |
AC005863.1 |
| chr6_-_167040731 | 0.05 |
ENST00000265678.4 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr6_-_128841503 | 0.05 |
ENST00000368215.3 ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
| chr2_-_70475701 | 0.05 |
ENST00000282574.4 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr7_-_107643674 | 0.05 |
ENST00000222399.6 |
LAMB1 |
laminin, beta 1 |
| chr17_+_26698677 | 0.05 |
ENST00000457710.3 |
SARM1 |
sterile alpha and TIR motif containing 1 |
| chr1_-_26232951 | 0.05 |
ENST00000426559.2 ENST00000455785.2 |
STMN1 |
stathmin 1 |
| chr10_+_104535994 | 0.05 |
ENST00000369889.4 |
WBP1L |
WW domain binding protein 1-like |
| chr7_+_107224364 | 0.05 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr6_-_75915757 | 0.05 |
ENST00000322507.8 |
COL12A1 |
collagen, type XII, alpha 1 |
| chr2_-_225266743 | 0.05 |
ENST00000409685.3 |
FAM124B |
family with sequence similarity 124B |
| chr6_+_126240442 | 0.05 |
ENST00000448104.1 ENST00000438495.1 ENST00000444128.1 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr12_-_49582978 | 0.04 |
ENST00000301071.7 |
TUBA1A |
tubulin, alpha 1a |
| chr17_-_49124230 | 0.04 |
ENST00000510283.1 ENST00000510855.1 |
SPAG9 |
sperm associated antigen 9 |
| chr16_+_14980632 | 0.04 |
ENST00000565655.1 |
NOMO1 |
NODAL modulator 1 |
| chr19_-_10047219 | 0.04 |
ENST00000264833.4 |
OLFM2 |
olfactomedin 2 |
| chr21_-_16437126 | 0.04 |
ENST00000318948.4 |
NRIP1 |
nuclear receptor interacting protein 1 |
| chr2_-_1748214 | 0.04 |
ENST00000433670.1 ENST00000425171.1 ENST00000252804.4 |
PXDN |
peroxidasin homolog (Drosophila) |
| chr1_+_244998602 | 0.04 |
ENST00000411948.2 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
| chr1_+_172502336 | 0.04 |
ENST00000263688.3 |
SUCO |
SUN domain containing ossification factor |
| chr12_+_60058458 | 0.04 |
ENST00000548610.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_+_46926326 | 0.04 |
ENST00000394861.2 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr12_+_124997766 | 0.04 |
ENST00000543970.1 |
RP11-83B20.1 |
RP11-83B20.1 |
| chr17_+_9066252 | 0.04 |
ENST00000436734.1 |
NTN1 |
netrin 1 |
| chr3_+_39509070 | 0.04 |
ENST00000354668.4 ENST00000428261.1 ENST00000420739.1 ENST00000415443.1 ENST00000447324.1 ENST00000383754.3 |
MOBP |
myelin-associated oligodendrocyte basic protein |
| chr19_-_18717627 | 0.04 |
ENST00000392386.3 |
CRLF1 |
cytokine receptor-like factor 1 |
| chr11_-_6677018 | 0.04 |
ENST00000299441.3 |
DCHS1 |
dachsous cadherin-related 1 |
| chr12_-_71003568 | 0.04 |
ENST00000547715.1 ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr3_-_167813672 | 0.04 |
ENST00000470487.1 |
GOLIM4 |
golgi integral membrane protein 4 |
| chr11_+_126225529 | 0.04 |
ENST00000227495.6 ENST00000444328.2 ENST00000356132.4 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr21_-_16437255 | 0.04 |
ENST00000400199.1 ENST00000400202.1 |
NRIP1 |
nuclear receptor interacting protein 1 |
| chr22_+_31160239 | 0.04 |
ENST00000445781.1 ENST00000401475.1 |
OSBP2 |
oxysterol binding protein 2 |
| chr1_-_26233423 | 0.04 |
ENST00000357865.2 |
STMN1 |
stathmin 1 |
| chr2_-_70475730 | 0.04 |
ENST00000445587.1 ENST00000433529.2 ENST00000415783.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr1_-_26232522 | 0.04 |
ENST00000399728.1 |
STMN1 |
stathmin 1 |
| chr12_+_64238539 | 0.04 |
ENST00000357825.3 |
SRGAP1 |
SLIT-ROBO Rho GTPase activating protein 1 |
| chr14_-_91884150 | 0.04 |
ENST00000553403.1 |
CCDC88C |
coiled-coil domain containing 88C |
| chr16_+_30406721 | 0.04 |
ENST00000320159.2 |
ZNF48 |
zinc finger protein 48 |
| chr3_-_149510553 | 0.04 |
ENST00000462519.2 ENST00000446160.1 ENST00000383050.3 |
ANKUB1 |
ankyrin repeat and ubiquitin domain containing 1 |
| chr16_-_15736881 | 0.04 |
ENST00000540441.2 |
KIAA0430 |
KIAA0430 |
| chr5_-_176900610 | 0.04 |
ENST00000477391.2 ENST00000393565.1 ENST00000309007.5 |
DBN1 |
drebrin 1 |
| chr7_-_82792215 | 0.04 |
ENST00000333891.9 ENST00000423517.2 |
PCLO |
piccolo presynaptic cytomatrix protein |
| chr1_-_1243392 | 0.04 |
ENST00000354700.5 |
ACAP3 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr2_+_65215604 | 0.04 |
ENST00000531327.1 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr11_+_131781290 | 0.04 |
ENST00000425719.2 ENST00000374784.1 |
NTM |
neurotrimin |
| chr6_-_3227877 | 0.04 |
ENST00000259818.7 |
TUBB2B |
tubulin, beta 2B class IIb |
| chr20_-_43150601 | 0.04 |
ENST00000541235.1 ENST00000255175.1 ENST00000342374.4 |
SERINC3 |
serine incorporator 3 |
| chr20_+_60718785 | 0.04 |
ENST00000421564.1 ENST00000450482.1 ENST00000331758.3 |
SS18L1 |
synovial sarcoma translocation gene on chromosome 18-like 1 |
| chr5_-_134871639 | 0.03 |
ENST00000314744.4 |
NEUROG1 |
neurogenin 1 |
| chr7_+_134464376 | 0.03 |
ENST00000454108.1 ENST00000361675.2 |
CALD1 |
caldesmon 1 |
| chr17_-_18585131 | 0.03 |
ENST00000443457.1 ENST00000583002.1 |
ZNF286B |
zinc finger protein 286B |
| chr16_+_54964740 | 0.03 |
ENST00000394636.4 |
IRX5 |
iroquois homeobox 5 |
| chr7_+_134464414 | 0.03 |
ENST00000361901.2 |
CALD1 |
caldesmon 1 |
| chr15_+_63335899 | 0.03 |
ENST00000561266.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr5_-_114515734 | 0.03 |
ENST00000514154.1 ENST00000282369.3 |
TRIM36 |
tripartite motif containing 36 |
| chrX_+_80457442 | 0.03 |
ENST00000373212.5 |
SH3BGRL |
SH3 domain binding glutamic acid-rich protein like |
| chr9_-_98269481 | 0.03 |
ENST00000418258.1 ENST00000553011.1 ENST00000551845.1 |
PTCH1 |
patched 1 |
| chr11_-_69519410 | 0.03 |
ENST00000294312.3 |
FGF19 |
fibroblast growth factor 19 |
| chr19_+_10713112 | 0.03 |
ENST00000590382.1 ENST00000407327.4 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
| chr1_+_249200395 | 0.03 |
ENST00000355360.4 ENST00000329291.5 ENST00000539153.1 |
PGBD2 |
piggyBac transposable element derived 2 |
| chr5_+_134094461 | 0.03 |
ENST00000452510.2 ENST00000354283.4 |
DDX46 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr4_+_95129061 | 0.03 |
ENST00000354268.4 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_26585686 | 0.03 |
ENST00000505206.1 ENST00000511789.1 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr7_+_116312411 | 0.03 |
ENST00000456159.1 ENST00000397752.3 ENST00000318493.6 |
MET |
met proto-oncogene |
| chr12_+_15475462 | 0.03 |
ENST00000543886.1 ENST00000348962.2 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr9_+_139221880 | 0.03 |
ENST00000392945.3 ENST00000440944.1 |
GPSM1 |
G-protein signaling modulator 1 |
| chrX_+_135579670 | 0.03 |
ENST00000218364.4 |
HTATSF1 |
HIV-1 Tat specific factor 1 |
| chr2_+_30569506 | 0.03 |
ENST00000421976.2 |
AC109642.1 |
AC109642.1 |
| chr3_-_18466026 | 0.03 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr1_-_26324534 | 0.03 |
ENST00000374284.1 ENST00000441420.1 ENST00000374282.3 |
PAFAH2 |
platelet-activating factor acetylhydrolase 2, 40kDa |
| chr1_-_161014731 | 0.03 |
ENST00000368020.1 |
USF1 |
upstream transcription factor 1 |
| chr9_-_123476719 | 0.03 |
ENST00000373930.3 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr17_-_62308087 | 0.03 |
ENST00000583097.1 |
TEX2 |
testis expressed 2 |
| chr12_+_93096619 | 0.03 |
ENST00000397833.3 |
C12orf74 |
chromosome 12 open reading frame 74 |
| chr7_+_87563458 | 0.03 |
ENST00000398204.4 |
ADAM22 |
ADAM metallopeptidase domain 22 |
| chr1_+_150229554 | 0.03 |
ENST00000369111.4 |
CA14 |
carbonic anhydrase XIV |
| chr7_-_128415844 | 0.03 |
ENST00000249389.2 |
OPN1SW |
opsin 1 (cone pigments), short-wave-sensitive |
| chr14_-_106926724 | 0.03 |
ENST00000434710.1 |
IGHV3-43 |
immunoglobulin heavy variable 3-43 |
| chr4_+_95128996 | 0.03 |
ENST00000457823.2 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chrX_-_106959631 | 0.03 |
ENST00000486554.1 ENST00000372390.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr4_-_56458374 | 0.03 |
ENST00000295645.4 |
PDCL2 |
phosducin-like 2 |
| chrX_-_153599578 | 0.03 |
ENST00000360319.4 ENST00000344736.4 |
FLNA |
filamin A, alpha |
| chr6_-_35464817 | 0.03 |
ENST00000338863.7 |
TEAD3 |
TEA domain family member 3 |
| chr9_-_98269699 | 0.03 |
ENST00000429896.2 |
PTCH1 |
patched 1 |
| chr13_-_41593425 | 0.03 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr14_-_96830207 | 0.03 |
ENST00000359933.4 |
ATG2B |
autophagy related 2B |
| chr12_-_50294033 | 0.03 |
ENST00000552669.1 |
FAIM2 |
Fas apoptotic inhibitory molecule 2 |
| chr9_+_4662282 | 0.03 |
ENST00000381883.2 |
PPAPDC2 |
phosphatidic acid phosphatase type 2 domain containing 2 |
| chr6_+_83777374 | 0.03 |
ENST00000349129.2 ENST00000237163.5 ENST00000536812.1 |
DOPEY1 |
dopey family member 1 |
| chr13_+_24144509 | 0.03 |
ENST00000248484.4 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr3_-_65583561 | 0.03 |
ENST00000460329.2 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr11_-_69633792 | 0.03 |
ENST00000334134.2 |
FGF3 |
fibroblast growth factor 3 |
| chr19_-_44124019 | 0.03 |
ENST00000300811.3 |
ZNF428 |
zinc finger protein 428 |
| chr9_+_124103625 | 0.03 |
ENST00000594963.1 |
AL161784.1 |
Uncharacterized protein |
| chr12_+_93096759 | 0.03 |
ENST00000544406.2 |
C12orf74 |
chromosome 12 open reading frame 74 |
| chr12_+_104359641 | 0.03 |
ENST00000537100.1 |
TDG |
thymine-DNA glycosylase |
| chr1_-_154155675 | 0.03 |
ENST00000330188.9 ENST00000341485.5 |
TPM3 |
tropomyosin 3 |
| chr5_+_49962772 | 0.03 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr4_-_186877502 | 0.03 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr5_-_146435501 | 0.03 |
ENST00000336640.6 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr19_-_17571613 | 0.03 |
ENST00000594663.1 |
CTD-2521M24.10 |
Uncharacterized protein |
| chr9_-_39239171 | 0.03 |
ENST00000358144.2 |
CNTNAP3 |
contactin associated protein-like 3 |
| chr17_-_53046058 | 0.03 |
ENST00000571584.1 ENST00000299335.3 |
COX11 |
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr7_+_87563557 | 0.03 |
ENST00000439864.1 ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22 |
ADAM metallopeptidase domain 22 |
| chr6_-_35464727 | 0.03 |
ENST00000402886.3 |
TEAD3 |
TEA domain family member 3 |
| chr17_-_56065484 | 0.03 |
ENST00000581208.1 |
VEZF1 |
vascular endothelial zinc finger 1 |
| chr5_-_146435694 | 0.03 |
ENST00000356826.3 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr1_+_201857798 | 0.03 |
ENST00000362011.6 |
SHISA4 |
shisa family member 4 |
| chr2_+_54683419 | 0.03 |
ENST00000356805.4 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
| chr4_+_95128748 | 0.02 |
ENST00000359052.4 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr12_-_105478339 | 0.02 |
ENST00000424857.2 ENST00000258494.9 |
ALDH1L2 |
aldehyde dehydrogenase 1 family, member L2 |
| chr2_-_2334888 | 0.02 |
ENST00000428368.2 ENST00000399161.2 |
MYT1L |
myelin transcription factor 1-like |
| chrX_+_12993202 | 0.02 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr14_-_35344093 | 0.02 |
ENST00000382422.2 |
BAZ1A |
bromodomain adjacent to zinc finger domain, 1A |
| chr2_-_55276320 | 0.02 |
ENST00000357376.3 |
RTN4 |
reticulon 4 |
| chr3_+_39509163 | 0.02 |
ENST00000436143.2 ENST00000441980.2 ENST00000311042.6 |
MOBP |
myelin-associated oligodendrocyte basic protein |
| chr15_-_56035177 | 0.02 |
ENST00000389286.4 ENST00000561292.1 |
PRTG |
protogenin |
| chr20_-_39317868 | 0.02 |
ENST00000373313.2 |
MAFB |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr8_+_126442563 | 0.02 |
ENST00000311922.3 |
TRIB1 |
tribbles pseudokinase 1 |
| chr21_+_33784670 | 0.02 |
ENST00000300255.2 |
EVA1C |
eva-1 homolog C (C. elegans) |
| chr2_-_128568721 | 0.02 |
ENST00000322313.4 ENST00000393006.1 ENST00000409658.3 ENST00000436787.1 |
WDR33 |
WD repeat domain 33 |
| chr20_+_30555805 | 0.02 |
ENST00000562532.2 |
XKR7 |
XK, Kell blood group complex subunit-related family, member 7 |
| chr1_+_200993071 | 0.02 |
ENST00000446333.1 ENST00000458003.1 |
RP11-168O16.1 |
RP11-168O16.1 |
| chr7_+_102553430 | 0.02 |
ENST00000339431.4 ENST00000249377.4 |
LRRC17 |
leucine rich repeat containing 17 |
| chr2_-_70475586 | 0.02 |
ENST00000416149.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr6_+_47666275 | 0.02 |
ENST00000327753.3 ENST00000283303.2 |
GPR115 |
G protein-coupled receptor 115 |
| chr5_-_146435572 | 0.02 |
ENST00000394414.1 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr6_+_13272904 | 0.02 |
ENST00000379335.3 ENST00000379329.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.1 | GO:0038162 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.0 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.0 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0039714 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.0 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.0 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |